Milestones in Cancer Research and Discovery
During the past 250 years, we have witnessed many landmark discoveries in our efforts to make progress against cancer, an affliction known to humanity for thousands of years. This timeline shows a few key milestones in the history of cancer research.
1775: Chimney Soot & Squamous Cell Carcinoma
Percivall Pott identifies a relationship between exposure to chimney soot and the incidence of squamous cell carcinoma of the scrotum among chimney sweeps. His report is the first to clearly link an environmental exposure to the development of cancer.
1863: Inflammation & Cancer
Rudolph Virchow identifies white blood cells (leukocytes) in cancerous tissue, making the first connection between inflammation and cancer. Virchow also coins the term "leukemia" and is the first person to describe the excess number of white blood cells in the blood of patients with this disease.
1882: The First Radical Mastectomy to Treat Breast Cancer
William Halsted performs the first radical mastectomy to treat breast cancer. This surgical procedure remains the standard operation for breast cancer until the latter half of the 20th century.
1886: Inheritance of Cancer Risk
Brazilian ophthalmologist Hilário de Gouvêa provides the first documented evidence that a susceptibility to cancer can be passed on from a parent to a child. He reports that two of seven children born to a father who was successfully treated for childhood retinoblastoma, a malignant tumor of the eye, also developed the disease.
1895: The First X-Ray
Wilhelm Roentgen discovers x-rays. The first x-ray picture is an image of his wife's hand.
1898: Radium & Polonium
Marie and Pierre Curie discover the radioactive elements radium and polonium. Within a few years, the use of radium in cancer treatment begins.
1899: The First Use of Radiation Therapy to Cure Cancer
Swedish physicians Tor Stenbeck and Tage Sjogren describe the first cases of basal cell carcinoma of the skin and squamous cell carcinoma of the skin cured by X-ray therapy.
1902: Cancer Tumors & Single Cells with Chromosome Damage
Theodor Boveri proposes that cancerous tumors arise from single cells that have experienced chromosome damage and suggests that chromosome alterations cause the cells to divide uncontrollably.
1909: Immune Surveillance
Paul Ehrlich proposes that the immune system usually suppresses tumor formation, a concept that becomes known as the "immune surveillance" hypothesis. This proposal prompts research, which continues today, to harness the power of the immune system to fight cancer.
1911: Cancer in Chickens
Peyton Rous discovers a virus that causes cancer in chickens (Rous sarcoma virus), establishing that some cancers are caused by infectious agents.
1915: Cancer in Rabbits
Katsusaburo Yamagiwa and Koichi Ichikawa induce cancer in rabbits by applying coal tar to their skin, providing experimental proof that chemicals can cause cancer.
1928: The Pap Smear
George Papanicolaou discovers that cervical cancer can be detected by examining cells from the vagina under a microscope. This breakthrough leads to the development of the Pap test, which allows abnormal cervical cells to be detected and removed before they become cancerous.
1932: The Modified Radical Mastectomy for Breast Cancer
David H. Patey develops the modified radical mastectomy for breast cancer. This surgical procedure is less disfiguring than the radical mastectomy and eventually replaces it as the standard surgical treatment for breast cancer.
1937: The National Cancer Institute (NCI)
Legislation signed by President Franklin D. Roosevelt establishes the National Cancer Institute (NCI).
1937: Breast-Sparing Surgery Followed by Radiation
Sir Geoffrey Keynes describes the treatment of breast cancer with breast-sparing surgery followed by radiation therapy. After surgery to remove the tumor, long needles containing radium are inserted throughout the affected breast and near the adjacent axillary lymph nodes.
1941: Hormonal Therapy
Charles Huggins discovers that removing the testicles to lower testosterone production or administering estrogens causes prostate tumors to regress. Such hormonal manipulation—more commonly known as hormonal therapy—continues to be a mainstay of prostate cancer treatment.
1947: Antimetabolites
Sidney Farber shows that treatment with the antimetabolite drug aminopterin, a derivative of folic acid, induces temporary remissions in children with acute leukemia. Antimetabolite drugs are structurally similar to chemicals needed for important cellular processes, such as DNA synthesis, and cause cell death by blocking those processes.
1949: Nitrogen Mustard
The Food and Drug Administration (FDA) approves nitrogen mustard (mechlorethamine) for the treatment of cancer. Nitrogen mustard belongs to a class of drugs called alkylating agents, which kill cells by chemically modifying their DNA.
1950: Cigarette Smoking & Lung Cancer
Ernst Wynder, Evarts Graham, and Richard Doll identify cigarette smoking as an important factor in the development of lung cancer.
1953: The First Complete Cure of a Human Solid Tumor
Roy Hertz and Min Chiu Li achieve the first complete cure of a human solid tumor by chemotherapy when they use the drug methotrexate to treat a patient with choriocarcinoma, a rare cancer of the reproductive tissue that mainly affects women.
1958: Combination Chemotherapy
NCI researchers Emil Frei, Emil Freireich, and James Holland and their colleagues demonstrate that combination chemotherapy with the drugs 6-mercaptopurine and methotrexate can induce partial and complete remissions and prolong survival in children and adults with acute leukemia.
1960: The Philadelphia Chromosome
Peter Nowell and David Hungerford describe an unusually small chromosome in the cancer cells of patients with chronic myelogenous leukemia (CML). This chromosome, which becomes known as the Philadelphia chromosome, is found in the leukemia cells of 95% of patients with CML.
1964: A Focus on Cigarette Smoking
The U.S. Surgeon General issues a report stating that cigarette smoking is an important health hazard in the United States and that action is required to reduce its harmful effects.
1964: The Epstein-Barr virus
For the first time, a virus—the Epstein-Barr virus (EBV)—is linked to a human cancer (Burkitt lymphoma). EBV is later shown to cause several other cancers, including nasopharyngeal carcinoma, Hodgkin lymphoma, and some gastric (stomach) cancers.
1971: The National Cancer Act
On December 23, President Richard M. Nixon signs the National Cancer Act, which authorizes the NCI Director to coordinate all activities of the National Cancer Program, establish national cancer research centers, and establish national cancer control programs.
1976: The DNA of Normal Chicken Cells
Dominique Stehelin, Harold Varmus, J. Michael Bishop, and Peter Vogt discover that the DNA of normal chicken cells contains a gene related to the oncogene (cancer-causing gene) of avian sarcoma virus, which causes cancer in chickens. This finding eventually leads to the discovery of human oncogenes.
1978: Tamoxifen
FDA approves tamoxifen, an antiestrogen drug originally developed as a birth control treatment, for the treatment of breast cancer. Tamoxifen represents the first of a class of drugs known as selective estrogen receptor modulators, or SERMs, to be approved for cancer therapy.
1979: The TP53 Gene
The TP53 gene (also called p53), the most commonly mutated gene in human cancer, is discovered. It is a tumor suppressor gene, meaning its protein product (p53 protein) helps control cell proliferation and suppress tumor growth.
1984: HER2 Gene Discovered
Researchers discover a new oncogene in rat cells that they call “neu.” The human version of this gene, called HER2 (and ErbB2), is overexpressed in about 20% to 25% of breast cancers (known as HER2-positive breast cancers) and is associated with more aggressive disease and a poor prognosis.
1984: HPV 16 & 18
DNA from human papillomavirus (HPV) types 16 and 18 is identified in a large percentage of cervical cancers, establishing a link between infection with these HPV types and cervical carcinogenesis.
1985: Breast-Conserving Surgery
Results from an NCI-supported clinical trial show that women with early-stage breast cancer who were treated with breast-conserving surgery (lumpectomy) followed by whole-breast radiation therapy had similar rates of overall survival and disease-free survival as women who were treated with mastectomy alone.
1986: HER2 Oncogene Cloning
The human oncogene HER2 (also called neu and erbB2) is cloned. Overexpression of the protein product of this gene, which occurs in about 20% to 25% of breast cancers (known as HER2-positive breast cancers), is associated with more aggressive disease and a poor prognosis.
1993: Guaiac Fecal Occult Blood Testing (FOBT)
Results from an NCI-supported clinical trial show that annual screening with guaiac fecal occult blood testing (FOBT) can reduce colorectal cancer mortality by about 33%.
1994: BRCA1 Tumor Suppressor Gene Cloning
The tumor suppressor gene BRCA1 is cloned. Specific inherited mutations in this gene greatly increase the risks of breast and ovarian cancer in women and the risks of several other cancers in both men and women.
1995: BRCA2 Tumor Suppressor Gene Cloning
The tumor suppressor gene BRCA2 is cloned. Similar to BRCA1, inheriting specific BRCA2 gene mutations greatly increases the risks of breast and ovarian cancer in women and the risks of several other cancers in both men and women.
1996: Anastrozole
FDA approves anastrozole for the treatment of estrogen receptor-positive advanced breast cancer in postmenopausal women. Anastrozole is the first aromatase inhibitor (a drug that blocks the production of estrogen in the body) to be approved for cancer therapy.
1997: Rituximab
FDA approves rituximab, a monoclonal antibody, for use in patients with treatment-resistant, low-grade or follicular B-cell non-Hodgkin lymphoma (NHL). Rituximab is the first monoclonal antibody approved for use in cancer therapy. It is later approved as an initial treatment for these types of NHL, for another type of NHL called diffuse large B-cell lymphoma, and for chronic lymphocytic leukemia.
1998: NCI-Sponsored Breast Cancer Prevention Trial
Results of the NCI-sponsored Breast Cancer Prevention Trial show that the antiestrogen drug tamoxifen can reduce the incidence of breast cancer among women who are at increased risk of the disease by about 50%. FDA approves tamoxifen to reduce the incidence of breast cancer in women at increased risk.
1998: Trastuzumab
FDA approves trastuzumab, a monoclonal antibody that targets cancer cells that overexpress the HER2 gene, for the treatment of women with HER2-positive metastatic breast cancer. Trastuzumab is later approved for the adjuvant (post-operative) treatment of women with HER2-positive early-stage breast cancer.
2001: Imatinib Mesylate
Results of a clinical trial show that the drug imatinib mesylate, which targets a unique protein produced by the Philadelphia chromosome, is effective against chronic myelogenous leukemia (CML). Imatinib treatment changes the usually fatal disease into a manageable condition. Later, it is also shown to be effective in the treatment of gastrointestinal stromal tumors (GIST).
2003: NCI-Sponsored Prostate Cancer Prevention Trial (PCPT)
Results of the NCI-sponsored Prostate Cancer Prevention Trial (PCPT) show that the drug finasteride, which reduces the production of male hormones in the body, lowers a man's risk of prostate cancer by about 25%.
2006: NCI's Study of Tamoxifen and Raloxifene (STAR)
Results of NCI's Study of Tamoxifen and Raloxifene (STAR) show that postmenopausal women at increased risk of breast cancer can reduce their risk of developing the disease if they take the antiestrogen drug raloxifene. The risk of serious side effects is lower with raloxifene than with tamoxifen.
2006: Gardasil
FDA approves the human papillomavirus (HPV) vaccine Gardasil, which protects against infection by the two HPV types (HPV 16 and 18) that cause approximately 70% of all cases of cervical cancer and two additional HPV types (HPV 6 and 11) that cause 90% of genital warts. Gardasil is the first vaccine approved to prevent cervical cancer. NCI scientists made technological advances that enabled development of Gardasil and subsequent HPV vaccines.
2009: Cervarix
FDA approves Cervarix, a second vaccine that protects against infection by the two HPV types that cause approximately 70% of all cases of cervical cancer worldwide.
2010: The First Human Cancer Treatment Vaccine
FDA approves sipuleucel-T, a cancer treatment vaccine that is made using a patient's own immune system cells (dendritic cells), for the treatment of metastatic prostate cancer that no longer responds to hormonal therapy. It is the first (and so far only) human cancer treatment vaccine to be approved.
2010: NCI-Sponsored Lung Cancer Screening Trial (NLST)
Initial results of the NCI-sponsored Lung Cancer Screening Trial (NLST) show that screening with low-dose helical computerized tomography (CT) reduced lung cancer deaths by about 20% in a large group of current and former heavy smokers.
2011: Ipilimumab
FDA approves the use of ipilimumab, a monoclonal antibody, for the treatment of inoperable or metastatic melanoma. Ipilimumab stimulates the immune system to attack cancer cells by removing a "brake" that normally controls the intensity of immune responses.
2012: NCI-Sponsored PLCO Cancer Screening Trial
Results of the NCI-sponsored PLCO Cancer Screening Trial confirm that screening people 55 years of age and older for colorectal cancer using flexible sigmoidoscopy reduces colorectal cancer incidence and mortality. In the PLCO trial, screened individuals had a 21% lower risk of developing colorectal cancer and a 26% lower risk of dying from the disease than the control subjects.
2013: Ado-Trastuzumab Emtansine (T-DM1)
FDA approves ado-trastuzumab emtansine (T-DM1) for the treatment of patients with HER2-positive breast cancer who were previously treated with trastuzumab and/or a taxane drug. T-DM1 is an immunotoxin (an antibody-drug conjugate) that is made by chemically linking the monoclonal antibody trastuzumab to the cytotoxic agent mertansine, which inhibits cell proliferation by blocking the formation of microtubules.
2014: Analyzing DNA in Cancer
Researchers from The Cancer Genome Atlas (TCGA) project, a joint effort by NCI and the National Human Genome Research Institute to analyze the DNA and other molecular changes in more than 30 types of human cancer, find that gastric (stomach) cancer is actually four different diseases, not just one, based on differing tumor characteristics. This finding from TCGA and other related projects may potentially lead to a new classification system for cancer, in which cancers are classified by their molecular abnormalities as well as their organ or tissue site of origin.
2014: Pembrolizumab
FDA approves pembrolizumab for the treatment of advanced melanoma. This monoclonal antibody blocks the activity of a protein called PD1 on immune cells, which increases the strength of immune responses against cancer.
2014: Gardasil 9
FDA approves Gardasil 9, a vaccine that protects against infection with the same four HPV types as Gardasil plus five more cancer-causing HPV types that together account for nearly 90% of cervical cancers. It is now the only HPV vaccine available in the United States.
2015: NCI-MATCH Clinical Trial
NCI and the ECOG-ACRIN Cancer Research Group launch the NCI-MATCH (Molecular Analysis for Therapy Choice) clinical trial to test more than 20 drugs and drug combinations based on molecular analysis of tumors in people with cancer. The study is designed to determine whether targeted therapies for people whose tumors have specific gene mutations will be effective regardless of their cancer type.
2015: Talimogene Laherparepvec
FDA approves talimogene laherparepvec (T-VEC) for the treatment of some patients with metastatic melanoma that cannot be surgically removed. T-VEC, the first oncolytic virus approved for clinical use, works by infecting and killing tumor cells and stimulating an immune response against cancer cells throughout the body.
2016: Cancer Moonshot℠
Congress passes the 21st Century Cures Act, which provides funding for the Cancer Moonshot, a broad program to accelerate cancer research by investing in specific research initiatives that have the potential to transform cancer care, detection, and prevention.
2017: Pediatric MATCH
NCI and the Children’s Oncology Group launch Pediatric MATCH, an effort to extend molecular analysis and targeted treatment to children and adolescents with cancer. Like NCI-MATCH, Pediatric MATCH seeks to determine if treating tumors with molecularly targeted drugs based on the tumor’s genetic characteristics rather than the type of cancer or cancer site will be effective.
2017: CAR T-Cell Therapies
FDA approves tisagenlecleucel to treat a form of acute lymphoblastic leukemia in certain children and young adults. FDA subsequently approves axicabtagene ciloleucel for patients with large B-cell lymphomas whose cancer has progressed after receiving at least two prior treatment regimens. Both treatments are chimeric antigen receptor (CAR) T-cell therapies that are personalized for each patient. To create these therapies, T cells are removed from the patient, genetically altered to recognize cancer-specific antigens, grown to large numbers in the lab, and then infused back into the patient to stimulate their immune system to attack cancer cells.
2017: Tumor-Agnostic Approval for Pembrolizumab
FDA extends approval of pembrolizumab to treat metastatic and inoperable solid tumors that have certain genetic changes, wherever they occur in the body , that have progressed following prior treatment and that have no alternative treatment options. With this tissue-agnostic approval, pembrolizumab becomes the first cancer treatment based solely on the presence of a genetic feature in a tumor, rather than a person’s cancer type.
2017: Genomic Profiling Tests
FDA clears two products to test tumors for genetic changes that may make the tumors susceptible to treatment with FDA-approved molecularly targeted drugs. In November, FDA authorizes the MSK-IMPACT test developed and used by Memorial Sloan Kettering Cancer Center to analyze tumors for potentially actionable changes in 468 cancer-related genes. In December, FDA approves the FoundationOne CDx test, which evaluates genetic changes in 324 genes known to fuel cancer growth. The FoundationOne test serves as a companion diagnostic for several FDA-approved drugs targeting five common types of cancer.
2018: TCGA PanCancer Atlas
NIH-funded researchers with TCGA complete an in-depth genomic analysis of 33 cancer types. The PanCancer Atlas provides a detailed genomic analysis of molecular and clinical data from more than 10,000 tumors that gives cancer researchers an unprecedented understanding of how, where, and why tumors arise in humans.
2018: NCI-Sponsored TAILORx Clinical Trial
Results from the NCI-sponsored Trial Assigning IndividuaLized Options for Treatment (Rx), or TAILORx, clinical trial show that most women with early-stage breast cancer do not benefit from having chemotherapy after surgery. The trial used a molecular test that assesses the expression of 21 genes associated with breast cancer recurrence to assign women with early-stage, hormone receptor–positive, HER2-negative breast cancer that hasn’t spread to the lymph nodes to the most appropriate and effective post-operative treatment. It is one of the first trials to examine a way to personalize cancer treatment
2018: Larotrectinib
FDA approves larotrectinib, the first drug that targets tumors with NTRK gene fusions. The approval is for pediatric or adult patients with metastatic or inoperable solid tumors that have worsened after previous treatment anywhere in the body driven by an NTRK gene fusion without a known acquired resistance mutation. Larotrectinib is the second drug approved to treat cancer with specific molecular features regardless of where the cancer is located.
2020: International Pan-Cancer Analysis of Whole Genomes
A consortium of international researchers analyzes more than 2,600 whole genomes from 38 types of cancer and matching normal tissues to identify common patterns of molecular changes. The Pan-Cancer Analysis of Whole Genomes study, which used data collected by the International Cancer Genome Consortium and TCGA, uncovers the complex role that changes throughout the genome play in cancer development, growth, and spread. The study also extends genomic analyses of cancer beyond the protein-coding regions to the complete genetic composition of cells.
- Type 2 Diabetes
- Heart Disease
- Digestive Health
- Multiple Sclerosis
- COVID-19 Vaccines
- Occupational Therapy
- Healthy Aging
- Health Insurance
- Public Health
- Patient Rights
- Caregivers & Loved Ones
- End of Life Concerns
- Health News
- Thyroid Test Analyzer
- Doctor Discussion Guides
- Hemoglobin A1c Test Analyzer
- Lipid Test Analyzer
- Complete Blood Count (CBC) Analyzer
- What to Buy
- Editorial Process
- Meet Our Medical Expert Board

The History of Cancer: Discovery and Treatment
History of cancer.
- Modern Advances
Frequently Asked Questions
Cancer may have been “discovered” and written about thousands of years ago. However, the disease itself has actually existed since before the evolution of humans.
It was first documented in Egypt about 5,000 years ago. Since that time, people from cultures all over the world have written about the disease and its potential treatments.
This article will look at what we know about the history of cancer. It will also talk about how our understanding of what causes cancer and how it can be treated has changed over time.
- 3000 BCE : The world’s earliest known mention of cancer was found in a papyrus document from ancient Egypt. It described tumors found in the breast . The cancer was treated by destroying the tissue with a hot instrument called “the fire drill”—a technique we now call “cauterization.” Some writings have shown that the ancient Egyptians could distinguish between cancerous (malignant) and noncancerous (benign) tumors.
- 460 BCE : In ancient Greece, Hippocrates thought there were four fluids in the body that influenced health: blood, phlegm , yellow bile , and black bile. He believed that having too much black bile in a part of the body caused cancer. For the next 1,400 years, people believed cancer was caused by too much black bile.
- 1628 : William Harvey, physician to King James I of England, dissected animals and human cadavers to learn more about how the body worked. When he published a book about the circulatory system, it upended ancient ideas and opened the door for more research on the workings of the human body.
- 1761 : Giovanni Morgagni of Padua published a book based on hundreds of autopsies he had performed on former patients of his, looking at both their clinical symptoms in life and his postmortem observations of their organs. This laid the groundwork for modern autopsies to determine the cause of someone’s death.
- 1775: A British surgeon named Percivall Pott discovered that testicular cancer was common in chimney sweeps. This was the first time a cancer was connected to an environmental cause.
- 17th century : The discovery of the lymphatic system led to new ideas about cancer. The lymphatic system includes the tissues, vessels, and organs that move a substance called lymph around your body. Lymph is an important part of your immune system. When the lymphatic system was discovered, it brought about the possibility that problems in this part of the body could cause cancer. This idea was called the lymph theory. It replaced Hippocrates’ theory about black bile and cancer.
- 1838 : Johannes Mueller, a German pathologist, showed that cancer is made of cells, not lymph. Mueller’s student, physician Rudolf Virchow, figured out that all our cells—even cancerous ones—come from other cells. However, he thought cancer spread in the body “like a liquid.”
- 1860 : A German surgeon named Karl Thiersch was the first person to prove that cancer spread through malignant cells.
How Cancer Was Named
Although most people cite Hippocrates as the first person to use the word cancer, he actually used the Greek words karkinos and karkinoma when he wrote about tumors. These words were related to the Greek word for “crab” because Hippocrates thought the insides of the tumors looked like crabs.
The Roman physician Celsus was the first to translate the word into the Latin word “cancer.”
20th Century to Present Day
The 20th century was an exciting time in cancer research. Carcinogens, chemotherapy , radiation therapy, and better ways to diagnose cancer were all discovered in these years. Some of the most important discoveries of the 20th century include:
- 1915 : Katsusaburo Yamagiwa and Koichi Ichikawa at Tokyo University applied coal tar to the skin of rabbits, inducing cancer and showing that some substances are carcinogens or cancer-causing.
- 1962 : James Watson and Frances Crick won a Nobel Prize for discovering the chemical structure of DNA.
- 1970s : Scientists discover oncogenes and tumor suppressor genes.
- 1981: Japanese professor Takeshi Hirayama published the first research linking lung cancer to second-hand smoke.
- 1982: Baruch S. Blumberg helped develop a vaccine against hepatitis B, a cause of liver cancer.
- 1989: The first gene therapy cancer treatments began to evolve.
- 1994: Scientists discovered the BRCA1 gene. This was the first known gene found to predispose a person to developing breast or ovarian cancer.
- 1999: Jan Walboomers and Michele Manos found evidence implicating human papillomavirus (HPV) to 99.7% percent of cervical cancers.
Today, we are still learning more about cancer. We have found ways to prevent and treat some forms of cancer and even cure others. Clinical trials have allowed scientists to test new ways to find and treat cancer. Some of this century’s notable discoveries so far include:
- 2006: The first vaccine against the HPV virus was approved in the United States.
- 2009: Researchers find that immunotherapy improves cure rates for children with neuroblastoma.
- 2011: Low-dose computed tomography (CT) scans help reduce lung cancer deaths by finding early-stage cancer in high-risk people.
- 2016: Researchers find evidence that a type of gene therapy called (CAR) T can produce remission in some people with B-cell hematologic cancers.
- 2021: The OncoKB, a genetic variant database, was recognized by the FDA as a tool for predicting drug responses in people with cancer. This will help oncologists find the best individual treatments for people with specific types of cancer.
Humans have known about cancer for millennia, but our modern understanding of cancer has only developed in the past few centuries. New advancements are being made all the time, and huge leaps have been made in the last few decades alone. This bodes well for the future of cancer treatments and therapies.
A Word From Verywell
How we look at cancer and its treatments has significantly changed in the last few centuries. Even decades ago, we had limited treatment options and less research. Learning about cancer and treatment history can be interesting when seeing how far we’ve come in such a short time. With new research and discoveries occurring all the time, the future of cancer research is an exciting topic.
Cancer has been around since humanity began recording its history and likely existed even before that time. The oldest description of cancer originates from Egypt around 3000 BC in a text called the Edwin Smith Papyrus, which also describes the Egyptian process of tumor removal using a method of cauterization.
Cancer was treated throughout most of the 1800s using surgery to remove cancerous tumors and affected organs. The discovery of X-rays in 1895 by a physicist named Wilhelm Konrad Roentgen helped to diagnose cancer cases and helped pave the way for radiation therapy.
In 1838, a pathologist known as Johannes Müller showed that cancer cells are what make up cancer. Before this, it was believed that cancer was made up of lymph.
It was first treated by surgery, although early physicians realized that cancer often came back after surgery.
The German chemist Paul Ehrlich started working with drugs to treat infectious diseases in the early 1900s. He coined the term “chemotherapy” to describe the use of chemicals to treat disease. He wasn’t very optimistic about medicine to treat cancer, though.
Cancer is more common with age, and more people are living longer, increasing the risk of cancer. A better metric of progress is the cancer death rate, which is decreasing, indicating that we are developing better treatments for cancer.
Di Lonardo A, Nasi S, Pulciani S. Cancer: we should not forget the past . J Cancer . 2015;6(1):29-39. doi:10.7150/jca.10336
American Cancer Society. Understanding cancer causes: ancient times to present .
National Cancer Institute. Cancer: a historic perspective .
Bolli R. William Harvey and the discovery of the circulation of the blood: part II . Circ Res . 2019;124(9):1300-1302. doi:10.1161/CIRCRESAHA.119.314977
Ghosh SK. Giovanni Battista Morgagni (1682-1771): father of pathologic anatomy and pioneer of modern medicine . Anat Sci Int . 2017;92(3):305-312. doi:10.1007/s12565-016-0373-7
Walter E, Scott M. The life and work of Rudolf Virchow 1821-1902: "Cell theory, thrombosis and the sausage duel" . J Intensive Care Soc . 2017;18(3):234–235. doi:10.1177/1751143716663967
Faguet GB. A brief history of cancer: age-old milestones underlying our current knowledge database . Int J Cancer . 2015;136(9):2022-2236. doi:10.1002/ijc.29134
Iida K, Proctor RN. 'The industry must be inconspicuous': Japan Tobacco's corruption of science and health policy via the Smoking Research Foundation . Tob Control . 2018;27(e1):e3-e11. doi:10.1136/tobaccocontrol-2017-053971
Gerlich WH. Medical virology of hepatitis B: how it began and where we are now . Virol J . 2013;10(1):1-25. doi:10.1186/1743-422X-10-239
National Institutes of Health. Gene therapy turns 30 years old .
Takaoka M, Miki Y. BRCA1 gene: function and deficiency . Int J Clin Oncol . 2018;23(1):36-44. doi:10.1007/s10147-017-1182-2
Okunade KS. Human papillomavirus and cervical cancer . J Obstet Gynaecol . 2020;40(5):602-608. doi:10.1080/01443615.2019.1634030
National Cancer Institute. The HPV vaccine .
National Cancer Institute. Harnessing the power of our immune systems to treat neuroblastoma: discovery of Ch14.18 immunotherapy .
National Cancer Institute. Lung cancer screening saves lives: the National Lung Screening Trial .
Park JH, Geyer MB, Brentjens RJ. CD19-targeted CAR T-cell therapeutics for hematologic malignancies: interpreting clinical outcomes to date . Blood J Am Soc Hematol . 2016;127(26):3312-20. doi:10.1182/blood-2016-02-629063
Food and Drug Administration. FDA recognizes Memorial Sloan-Kettering database of molecular tumor marker information .
American Cancer Society. Understanding what cancer is: ancient time to present .
American Cancer Society: The Cancer Atlas. History of cancer .
American Cancer Society. History of cancer treatments: surgery.
Valent P, Groner B, Schumacher U, et al. Paul Ehrlich (1854-1915) and his contributions to the foundation and birth of translational medicine . J Innate Immun . 2016;8(2):111-120. doi:10.1159/000443526
National Cancer Institute. Cancer statistics.
By Jaime R. Herndon, MS, MPH Herndon is a freelance health/medical writer with a graduate certificate in science writing from Johns Hopkins University.

Handbook of Oncobiology: From Basic to Clinical Sciences pp 1–29 Cite as
History, Evolution, Milestones in Cancer Research and Treatment
- Indu Sharma ORCID: orcid.org/0000-0001-9846-9787 4 ,
- Anuradha Sharma ORCID: orcid.org/0000-0001-5975-1456 4 ,
- Reena Tomer ORCID: orcid.org/0000-0003-1133-763X 4 ,
- Neha Negi ORCID: orcid.org/0000-0002-1902-7081 4 &
- Ranbir Chander Sobti 5
- Living reference work entry
- First Online: 15 July 2023
66 Accesses
The historical findings of patients with cancer from ancient Egyptian and Greek civilizations support the millennium long medical history of cancer. However, the disease at that time was mostly treated with not so effective radical surgery and cautery, making death the ultimate outcome of cancer patients. Over the centuries, various breakthrough discoveries have not only reformed the cancer detection but also contributed to the development of more effective therapeutic approaches. The most significant of them was the unearthing of cytotoxic antitumor drugs and the inception of chemotherapy. Since then, an exponential progress has been witnessed over the time about new cancer drugs. Another revolution in the field of oncology was targeted therapy with the development of specific drugs for some molecular targets involved in vital neoplastic processes. Collectively, chemotherapy and targeted therapy have definitely enhanced not only the survival rate but also the quality of life of cancer patients. In present times, genetic engineering studies have amplified the further advancements of cancer biology by utilizing monoclonal antibodies and immune checkpoint inhibitors specifically for advanced or metastatic tumors. Hence, cancer research has continuously grown with an intend to develop newer and better therapeutic approaches for cancer. Most recent, artificial intelligence and precision medicine are certainly going to bring a new revolution in the field of medical oncology.
This is a preview of subscription content, log in via an institution .
Alverson DC, Krupinski EA, Erps KA, Rowe NS, Weinstein RS (2019) The third national telemedicine & telehealth service provider showcase conference. Telemed J E Health 25(4):332–340. https://doi.org/10.1089/tmj.2018.0096
American Cancer Society. www.cancer.org | 1.800.227.2345
Amiri-Kordestani L, Blumenthal GM, Xu QC, Zhang L, Tang SW, Ha L, Weinberg WC, Chi B, Candau-Chacon R, Hughes P, Russell AM, Miksinski SP, Chen XH, McGuinn WD, Palmby T, Schrieber SJ, Liu Q, Wang J, Song P, Mehrotra N et al (2014) FDA approval: ado-trastuzumab emtansine for the treatment of patients with HER2-positive metastatic breast cancer. Clin Cancer Res 20(17):4436–4441. https://doi.org/10.1158/1078-0432.CCR-14-0012
Article CAS PubMed Google Scholar
Andriole GL, Bostwick DG, Brawley OW, Gomella LG, Marberger M, Montorsi F, Pettaway, CA, Tammela TL, Teloken C, Tindall DJ, Somerville MC, Wilson TH, Fowler IL, Rittmaster RS, & REDUCE Study Group (2010) Effect of dutasteride on the risk of prostate cancer. N Engl J Med 362(13):1192–1202. https://doi.org/10.1056/NEJMoa0908127
Azike JE (2009) A review of the history, epidemiology and treatment of squamous cell carcinoma of the scrotum. Rare Tumors 1(1):47–49
Article Google Scholar
Bagnardi V, Blangiardo M, La Vecchia C, Corrao G (2001) A meta-analysis of alcohol drinking and cancer risk. Br J Cancer 85(11), 1700–1705. https://doi.org/10.1054/bjoc.2001.2140
Bakkalci D, Jia Y, Winter JR, Lewis JE, Taylor GS, Stagg HR (2020) Risk factors for Epstein Barr virus-associated cancers: a systematic review, critical appraisal, and mapping of the epidemiological evidence. J Glob Health 10(1):010405. https://doi.org/10.7189/jogh.10.010405
Article PubMed PubMed Central Google Scholar
Basu A, Kuziemsky C, de Araújo Novaes M, Kleber A, Sales F, Al-Shorbaji N, Flórez-Arango JF, Gogia SB, Ho K Hunter, I Iyengar, S John, O John, S Kulatunga G Rajput, VK Ranatunga P, Udayasankaran JG (2021) Telehealth and the COVID-19 Pandemic: International Perspectives and a Health Systems Framework for Telehealth Implementation to Support Critical Response. Yearb Med Inform 30(1):126–133. https://doi.org/10.1055/s-0041-1726484
Bayat Mokhtari R, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das B, Yeger H (2017) Combination therapy in combating cancer. Oncotarget 8(23):38022–38043. https://doi.org/10.18632/oncotarget.16723
Article PubMed Google Scholar
Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard CW 3rd, Cornelisse CJ, Devilee P, Devlin B (2000) Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science (New York, N.Y.) 287(5454):848–851. https://doi.org/10.1126/science.287.5454.848
Berger MF, Mardis ER (2018) The emerging clinical relevance of genomics in cancer medicine. Nature reviews. Clin Oncol 15(6):353–365. https://doi.org/10.1038/s41571-018-0002-6
Article CAS Google Scholar
Bergljung L (2005) Sir Geoffrey Keynes 1887–1982. Kirurgisk pionjär, medicinhistoriker, humanist [Sir Geoffrey Keynes 1887–1982. Surgical pioneer, medical historian, humanist]. Svensk medicinhistorisk tidskrift 9(1):147–153
PubMed Google Scholar
Blumberg BS, Larouzé B, London WT, Werner B, Hesser JE, Millman I, Saimot G, Payet M (1975) The relation of infection with the hepatitis B agent to primary hepatic carcinoma. Am J Pathol 81(3):669–682. PMID: 174434 PMCID: PMC2032339
Google Scholar
Burd EM (2003) Human papillomavirus and cervical cancer. Clin Microbiol Rev 16(1):1–17. https://doi.org/10.1128/CMR.16.1.1-17.2003
Article CAS PubMed PubMed Central Google Scholar
Burns MC, O’Donnell A, Puzanov I (2016) Pembrolizumab for the treatment of advanced melanoma. Expert Opin Orphan Drugs 4(8):867–873. https://doi.org/10.1080/21678707.2016.1191348
Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ (2003) Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults N Engl J Med 348(17):1625–1638. https://doi.org/10.1056
Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216):1061–1068. https://doi.org/10.1038/nature07385
Chang MH (2011) Hepatitis B virus and cancer prevention. Recent results in cancer research. Fortschritte der Krebsforschung. Progres dans les recherches sur le cancer 188:75–84. https://doi.org/10.1007/978-3-642-10858-7_6
Chen Y, Jia Y, Song W, Zhang L (2018) Therapeutic potential of nitrogen mustard based hybrid molecules. Front Pharmacol 9:1453
Cheng L, Wang Y, Du J (2020) Human papillomavirus vaccines: an updated review. Vaccines 8(3):391. https://doi.org/10.3390/vaccines8030391
Clarke MJ (1998) Ovarian ablation in breast cancer, 1896 to 1998: milestones along hierarchy of evidence from case report to Cochrane review. BMJ 317(7167):1246–1248. https://doi.org/10.1136/bmj.317.7167.1246
Colapietro A, Mancini A, D’Alessandro AM, Festuccia C (2019) Crocetin and crocin from saffron in cancer chemotherapy and chemoprevention. Anti-Cancer Agents Med Chem 19(1):38–47. https://doi.org/10.2174/1871520619666181231112453
Collins FS, Morgan M, Patrinos A (2003) The Human Genome Project: lessons from large-scale biology. Science (New York, NY) 300(5617):286–290. https://doi.org/10.1126/science.1084564
Connell PP, Hellman S (2009) Advances in radiotherapy and implications for the next century: a historical perspective. Cancer Res 69(2):383–392
Cornejo CM, Jambusaria-Pahlajani A, Willenbrink TJ, Schmults CD, Arron ST, Ruiz ES (2020) Field cancerization: treatment. J Am Acad Dermatol 83(3):719–730. https://doi.org/10.1016/j.jaad.2020.03.127
Cragg GM, Newman DJ (2013) Natural products: a continuing source of novel drug leads. Biochimica et biophysica acta 1830(6):3670–3695. https://doi.org/10.1016/j.bbagen.2013.02.008
Crawford ED (2004) Hormonal therapy in prostate cancer: historical approaches. Rev Urol 6(Suppl 7):S3–S11
PubMed PubMed Central Google Scholar
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462(7274):739–744. https://doi.org/10.1038/nature08617
DeVita VT Jr, Chu E (2008) A history of cancer chemotherapy. Cancer Res 68(21):8643–8653
Dillard RS, Hampton CM, Strauss JD, Ke Z, Altomara D, Guerrero-Ferreira RC, Kiss G, Wright ER (2018) Biological Applications at the Cutting Edge of Cryo-Electron Microscopy. Microscopy and microanalysis: Microsc Microanal 24(4):406–419. https://doi.org/10.1017/S1431927618012382
Dotan E, Aggarwal C, Smith MR (2010) Impact of rituximab (Rituxan) on the treatment of B-cell non-Hodgkin’s lymphoma. P & T 35(3):148–157
D’Souza G, Kreimer AR, Viscidi R et al (2007) Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med 356:1944–1956
Dunn DB (2020) Larotrectinib and entrectinib: TRK inhibitors for the treatment of pediatric and adult patients with NTRK gene fusion. J Adv Pract Oncol 11(4):418–423. https://doi.org/10.6004/jadpro.2020.11.4.9
Elemento O (2021) The road from Rous sarcoma virus to precision medicine. J Exp Med 218(4)
Elion GB, Singer S, Hitchings GH (1954) Antagonists of nucleic acid derivatives. VIII. Synergism in combinations of biochemically related antimetabolites. J Biol Chem 208:477–488
Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N (1998) Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst 90(18):1371–1388. https://doi.org/10.1093/jnci/90.18.1371
Fröman N (1996) Marie and Pierre Curie and the discovery of polonium and radium. Palestra na royal swedish academy of sciences, em Estocolmo, Suécia
Gilchrest BA (2021) Actinic keratoses: reconciling the biology of field cancerization with treatment paradigms. J Investig Dermatol 141(4):727–731. https://doi.org/10.1016/j.jid.2020.09.002
Gompel A (2019) Hormones et cancers du sein [Hormone and breast cancer]. Presse medicale (Paris, France: 1983) 48(10):1085–1091. https://doi.org/10.1016/j.lpm.2019.09.021
Graziani G, Tentori L, Navarra P (2012) Ipilimumab: a novel immunostimulatory monoclonal antibody for the treatment of cancer. Pharmacol Res 65(1):9–22. https://doi.org/10.1016/j.phrs.2011.09.002
Graziani G, Lisi L, Tentori L, Navarra P (2022) Monoclonal Antibodies to CTLA-4 with Focus on Ipilimumab. Exp Suppl 113:295–350. https://doi.org/10.1007/978-3-030-91311-3_10
Guallar-Garrido S, Julián E (2020) Bacillus Calmette-Guérin (BCG) therapy for bladder cancer: an update. ImmunoTargets Ther 9:1–11. https://doi.org/10.2147/ITT.S202006
Gutierrez C, Schiff R (2011) HER2: biology, detection, and clinical implications. Arch Pathol Lab Med 135(1):55–62. https://doi.org/10.5858/2010-0454-RAR.1
Hammerstrom AE, Cauley DH, Atkinson BJ, Sharma P (2011) Cancer immunotherapy: sipuleucel-T and beyond. Pharmacotherapy 31(8):813–828. https://doi.org/10.1592/phco.31.8.813
Hansford S, Huntsman DG (2014) Boveri at 100: Theodor Boveri and genetic predisposition to cancer. J Pathol 234(2):142–145. https://doi.org/10.1002/path.4414
Hansson N, Moll F, Schultheiss D, Krischel M (2016) Remembering Charles B. Huggins’ Nobel Prize for Hormonal Treatment of Prostatic Cancer at its 50th Anniversary. Eur Uro 69(6):971–972. https://doi.org/10.1016/j.eururo.2016.01.030
Hitchings GH, Elion GB (1954) The chemistry and biochemistry of purine analogs. Ann NY Acad Sci 60:195–199
Hodi FS, Chesney J, Pavlick AC, Robert C, Grossmann KF, McDermott DF, Linette GP, Meyer N, Giguere JK, Agarwala SS, Shaheen M, Ernstoff MS, Minor DR, Salama AK, Taylor MH, Ott PA, Horak C, Gagnier P, Jiang J, Wolchok JD, Postow MA (2016) Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2-year overall survival outcomes in amulticentre, randomised, controlled, phase 2 trial. Lancet Oncol 17(11):1558–1568. https://doi.org/10.1016/S1470-2045(16)30366-7
Howard NP, Troggio M, Durel CE, Muranty H, Denancé C, Bianco L, Tillman J, van de Weg E (2021) Integration of Infinium and Axiom SNP array data in the outcrossing species Malus × domestica and causes for seemingly incompatible calls. BMC genomics 22(1):246. https://doi.org/10.1186/s12864-021-07565-7
Howard KK, Makki H, Novotny NM, Mi M, Nguyen N (2022) Value of robotic surgery simulation for training surgical residents and attendings: a systematic review protocol. BMJ open 12(6):e059439. https://doi.org/10.1136/bmjopen-2021-059439
Hunter B, Hindocha S, Lee RW (2022) The Role of Artificial Intelligence in Early Cancer Diagnosis. Cancers 14(6):1524. https://doi.org/10.3390/cancers14061524
Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F (2004) Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 350(23):2335–342. https://doi.org/10.1056/NEJMoa032691
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (2020) Pan-cancer analysis of whole genomes. Nature 578(7793):82–93. https://doi.org/10.1038/s41586-020-1969-6
Iqbal N, Iqbal N (2014a) Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol Biol Int 2014:852748. https://doi.org/10.1155/2014/852748
Iqbal N, Iqbal N (2014b) Imatinib: a breakthrough of targeted therapy in cancer. Chemother Res Pract 2014:357027. https://doi.org/10.1155/2014/357027
Jensen G (2010) Cryo-EM, Part C: analyses, interpretation, and case studies Academic Press
Yang JE, Larson MR, Sibert BS, Shrum, S, Wright ER (2021) CorRelator: Interactive software for real-time high precision cryo-correlative light and electron microscopy. J Struct Biol 213(2):107709. https://doi.org/10.1016/j.jsb.2021.107709
Jeyakumar A, Younis T (2012) Trastuzumab for HER2-positive metastatic breast cancer: clinical and economic considerations. Clinical medicine insights. Oncology 6:179–187. https://doi.org/10.4137/CMO.S6460
Jones OT, Calanzani N, Saji S, Duffy SW, Emery J, Hamilton W, Singh H, de Wit NJ, Walter FM (2021) Artificial Intelligence Techniques That May Be Applied to Primary Care Data to Facilitate Earlier Diagnosis of Cancer: Systematic Review. J Med Internet Res 23(3):23483. https://doi.org/10.2196/23483
Jordan VC (2014) Tamoxifen as the first targeted long-term adjuvant therapy for breast cancer. Endocr Relat Cancer 21(3):R235–R246. https://doi.org/10.1530/ERC-14-0092
Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, Wang YC, Pazdur R, FDA (2005) Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clinical cancer research : Clin Cancer Res 11(10): 3604–3608. https://doi.org/10.1158/1078-0432.CCR-04-2135
Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, Romano A (2017) Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci 18(2):308. https://doi.org/10.3390/ijms18020308
Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, Iken M, Vucur M, Weiss S, Heikenwalder M, Khan S, Gil J, Bruder D, Manns M, Schirmacher P, Tacke F, Zender L (2011) Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479(7374):547–551. https://doi.org/10.1038/nature10599
Kaufman KD, Dawber RP (1999) Finasteride, a Type 2 5alpha-reductase inhibitor, in the treatment of men with androgenetic alopecia. Expert Opin Investig Drugs 8(4):403–415. https://doi.org/10.1517/13543784.8.4.403
Khatami M (2018) Cancer; an induced disease of twentieth century! Induction of tolerance, increased entropy and ‘Dark Energy’: loss of biorhythms (Anabolism v. Catabolism). Clin Trans Med 7:20. https://doi.org/10.1186/s40169-018-0193-6
Kramer BS, Berg CD, Aberle DR, Prorok PC (2011) Lung cancer screening with low-dose helical CT: results from the National Lung Screening Trial (NLST). J Med Screen 18(3):109–111. https://doi.org/10.1258/jms.2011.011055
Lamm DL, Blumenstein BA, Crawford ED et al (1991) A randomized trial of intravesical doxorubicin and immunotherapy with bacille Calmette-Guerin for transitional-cell carcinoma of the bladder. N Engl J Med 325:1205–1209
Langner E, Rzeski W (2012) Dietary derived compounds in cancer chemoprevention. Contemp Oncol (Poznan, Poland) 16(5):394–400. https://doi.org/10.5114/wo.2012.31767
Lechner M, Liu J, Masterson L, Fenton TR (2022) HPV-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol 19(5):306–327. https://doi.org/10.1038/s41571-022-00603-7
Li MC, Hertz R, Bergenstal DM (1958) Therapy of choriocarcinoma and related trophoblastic tumors with folic acid and purine antagonists. N Engl J Med 259:66–74
Lin R, Tripuraneni P (2011) Radiation therapy in early-stage invasive breast cancer. Indian J Surg Oncol 2(2):101–111. https://doi.org/10.1007/s13193-011-0048-8
Liu C-J, Chen P-J (2020) Elimination of Hepatitis B in highly endemic settings: lessons learned in Taiwan and challenges ahead. Viruses 12(8):815. https://doi.org/10.3390/v12080815
Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B (2017) The different mechanisms of cancer drug resistance: a brief review. Adv Pharmaceut Bull 7(3):339–348. https://doi.org/10.15171/apb.2017.041
Milani M, Jha G, Potter DA (2009) Anastrozole use in early stage breast cancer of post-menopausal women. Clin Med Ther 1:141–156. https://doi.org/10.4137/cmt.s9
Milani A, Basirnejad M, Shahbazi S, Bolhassani A (2017) Carotenoids: biochemistry, pharmacology and treatment. Br J Pharmacol 174(11):1290–1324. https://doi.org/10.1111/bph.13625
Miller EA, Pinsky PF, Schoen RE, Prorok PC, Church TR (2019) Effect of flexible sigmoidoscopy screening on colorectal cancer incidence and mortality: long-term follow-up of the randomised US PLCO cancer screening trial. Lancet Gastroenterol Hepatol 4(2):101–110. https://doi.org/10.1016/S2468-1253(18)30358-3
Monie A, Hung CF, Roden R, Wu TC (2008) Cervarix: a vaccine for the prevention of HPV 16, 18-associated cervical cancer. Biol Targets Ther 2(1):97–105
Monteiro AN, Waizbort R (2007) The accidental cancer geneticist: hilario de gouvea and hereditary retinoblastoma. Cancer Biol Ther 6(5):811–813
Nadel MR, Berkowitz Z, Klabunde CN, Smith RA, Coughlin SS, White MC (2010) Fecal occult blood testing beliefs and practices of U.S. primary care physicians: serious deviations from evidence-based recommendations. J Gen Intern Med 25(8):833–839. https://doi.org/10.1007/s11606-010-1328-7
Newman DJ, Cragg GM (2020) Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 83(3):770–803. https://doi.org/10.1021/acs.jnatprod.9b01285
Nowell PC (2007) Discovery of the Philadelphia chromosome: a personal perspective. J Clin Investig 117(8):2033–2035. https://doi.org/10.1172/JCI31771
Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH (2013) Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493(7433):542–546. https://doi.org/10.1038/nature11743
Marcus L, Lemery SJ, Keegan P, Pazdur R (2019) FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clinical cancer research : Clin Cancer Res 25(13):3753–3758. https://doi.org/10.1158/1078-0432.CCR-18-4070
Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ (2015) Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science (New York, N.Y.) 348(6237):880–886. https://doi.org/10.1126/science.aaa6806
McLaughlin JR, Risch HA, Lubinski J, Moller P, Ghadirian P, Lynch H, Karlan B, Fishman D, Rosen B, Neuhausen SL, Offit K, Kauff N, Domchek S, Tung N, Friedman E, Foulkes W, Sun P, Narod SA, Hereditary Ovarian Cancer Clinical Study Group (2007) Reproductive risk factors for ovarian cancer in carriers of BRCA1 or BRCA2 mutations: a case-control study. Lancet Oncol 8(1):26–34. https://doi.org/10.1016/S1470-2045(06)70983-4
Medical Advisory Secretariat (2010) Robotic-assisted minimally invasive surgery for gynecologic and urologic oncology: an evidence-based analysis. Ont Health Technol Assess Ser 10(27):1–118 PMID: 23074405 PMCID: PMC3382308
Mizuki H, Shimoyama Y, Ishikawa T, Sasaki M (2022) A genomic sequence of the type II-A clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system in Mycoplasma salivarium strain ATCC 29803. J Oral Microbiol 14(1):2008153. https://doi.org/10.1080/20002297.2021.2008153
Panahi B, Majidi M, Hejazi MA (2022) Genome mining approach reveals the occurrence and diversity pattern of clustered regularly interspaced short palindromic repeats/CRISPR-associated systems in lactobacillus brevis strains. Front Microbiol 13:911706. https://doi.org/10.3389/fmicb.2022.911706
Paul A, Paul S (2014) The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Frontiers in bioscience (Landmark edition) 19(4):605–618. https://doi.org/10.2741/4230
Pérez-Herrero E, Fernández-Medarde A (2015) Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur J Pharmaceut Biopharmaceut 93:52–79. https://doi.org/10.1016/j.ejpb.2015.03.018
Plesca M, Bordea C, El Houcheimi B, Ichim E, Blidaru A (2016) Evolution of radical mastectomy for breast cancer. J Med Life 9(2):183
CAS PubMed PubMed Central Google Scholar
Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, Liedtke M, Rosenblatt J, Maus MV, Turka A, Lam LP, Morgan RA, Friedman K, Massaro M, Wang J, Russotti G, Yang Z, Campbell T, Hege K, Petrocca F, Kochenderfer JN (2019) Anti-BCMA CAR T-Cell Therapy bb2121 in Relapsed or Refractory Multiple Myeloma. N Engl J Med 380(18):1726–1737. https://doi.org/10.1056/NEJMoa1817226
Ramalingam SS, Khuri FR (2021) The National Cancer Act of 1971: a seminal milestone in the fight against cancer. Cancer 127(24):4532–4533. https://doi.org/10.1002/cncr.34001
Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R (2012) Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumorcells. Nat Biotechnol 30(8):777–782. https://doi.org/10.1038/nbt.2282
Redman MW, Tangen CM, Goodman PJ, Lucia MS, Coltman CA Jr, Thompson IM (2008) Finasteride does not increase the risk of high-grade prostate cancer: a bias-adjusted modeling approach. Cancer Prev Res (Philadelphia, PA) 1:174–181
Rivlin N, Brosh R, Oren M, Rotter V (2011) Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2(4):466–474. https://doi.org/10.1177/1947601911408889
Rock CL, Thomson CA, Sullivan KR, Howe CL, Kushi LH, Caan BJ, Neuhouser ML, Bandera EV, Wang Y, Robien K, Basen-Engquist KM, Brown JC, Courneya KS, Crane TE, Garcia DO, Grant BL, Hamilton KK, Hartman SJ, Kenfield SA, Martinez ME et al (2022) American Cancer Society nutrition and physical activity guideline for cancer survivors. CA 72(3):230–262. https://doi.org/10.3322/caac.21719
Samadi AK, Bilsland A, Georgakilas AG, Amedei A, Amin A, Bishayee A et al (2015) A multi-targeted approach to suppress tumor-promoting inflammation. In: Semin Cancer Biol, vol 35. Academic, pp S151–S184
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244):262–265. https://doi.org/10.1038/nature07935
Schrijver LH, Olsson H, Phillips KA, Terry MB, Goldgar DE, Kast K, Engel C, Mooij TM, Adlard J, Barrowdale D, Davidson R, Eeles R, Ellis S, Evans DG, Frost D, Izatt L, Porteous ME, Side LE, Walker L, Berthet P et al (2018) Oral contraceptive use and breast cancer risk: retrospective and prospective analyses from a BRCA1 and BRCA2 mutation carrier Cohort Study. JNCI Cancer Spect 2(2):pky023. https://doi.org/10.1093/jncics/pky023
Slaughter DP, Southwick HW, Smejkal W (1953) Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6(5):963–968. https://doi.org/10.1002/10970142(195309)6:5-963::aid-cncr2820060515>3.0.co;2-q
Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, Curtis C (2015) A Big Bang model of humancolorectal tumor growth. Nat Genet 47(3):209–216. https://doi.org/10.1038/ng.3214
Spain PD, Kadan-Lottick N (2012) Observations of unprecedented remissions following novel treatment for acute leukemia in children in 1948. J R Soc Med 105(4):177–181. https://doi.org/10.1258/jrsm.2012.12k013
Sporn MB, Dunlop NM, Newton DL, Smith JM (1976) Prevention of chemical carcinogenesis by vitamin A and its synthetic analogs (retinoids). Fed Proc 35(6):1332–1338. PMID: 770206
Stehelin D, Varmus HE, Bishop JM, Vogt PK (1976) DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260(5547):170–173. https://doi.org/10.1038/260170a0
Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, DeAngelo DJ, Levine RL, Flinn IW, Kantarjian HM, Collins R, Patel MR, Frankel AE, Stein A, Sekeres MA, Swords RT, Medeiros BC, Willekens C, Vyas P, Tallman MS (2017) Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130(6):722–731. https://doi.org/10.1182/blood-2017-04-779405
Stellman SD (2006) Ernst Wynder: a remembrance. Prev Med 43(4):239–245. https://doi.org/10.1016/j.ypmed.2006.08.007
Tacklind J, Fink HA, Macdonald R (2010) Finasteride for benign prostatic hyperplasia. Cochrane Database Syst Rev 10:601–615
Tan SY, Tatsumura Y (2015) George Papanicolaou (1883–1962): discoverer of the Pap smear. Singap Med J 56(10):586–587. https://doi.org/10.11622/smedj.2015155
Teh AL, Pan H, Lin X, Lim YI, Patro CP, Cheong CY, Gong M, MacIsaac JL, Kwoh CK, Meaney MJ, Kobor MS, Chong YS, Gluckman PD, Holbrook JD, Karnani N (2016) Comparison of Methyl-capture Sequencing vs. Infinium 450K methylation array for methylome analysis in clinical samples. Epigenetics 11(1):36–48. https://doi.org/10.1080/15592294.2015.1132136
Thompson IM, Klein EA, Lippman SM, Coltman CA, Djavan B (2003) Prevention of prostate cancer with finasteride: US/European perspective. Eur Urol 44(6):650–655. https://doi.org/10.1016/j.eururo.2003.11.001
Tian J, Lai D, Zhou L (2017) Secondary metabolites from Acremonium fungi: diverse structures and bioactivities. Mini Rev Med Chem 17(7):603–632. https://doi.org/10.2174/1389557516666160914194134
Timbang MR, Sim MW, Bewley AF, Farwell DG, Mantravadi A, Moore MG (2019) HPV-related oropharyngeal cancer: a review on burden of the disease and opportunities for prevention and early detection. Hum Vaccin Immunother 15(7–8):1920–1928. https://doi.org/10.1080/21645515.2019.1600985
Timoneda J, Rodríguez-Fernández L, Zaragozá R, Marín MP, Cabezuelo MT, Torres L, Viña JR, Barber T (2018) Vitamin A deficiency and the lung. Nutrients 10(9):1132. https://doi.org/10.3390/nu10091132
Tubiana M (1996) Wilhelm Conrad Röntgen and the discovery of X-rays. Bulletin de l’Académie nationale de médecine 180(1):97–108
CAS PubMed Google Scholar
Ullah MF, Usmani S, Shah A, Abuduhier FM (2022) Dietary molecules and experimental evidence of epigenetic influence in cancer chemoprevention: an insight. Semin Cancer Biol 83:319–334. https://doi.org/10.1016/j.semcancer.2020.10.011
Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, Bevers TB, Fehrenbacher L, Pajon ER Jr, Wade JL 3rd, Robidoux A, Margolese RG, James J, Lippman SM, Runowicz CD, Ganz PA, Reis SE, McCaskill-Stevens W, Ford LG, Jordan VC et al (2006) Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: the NSABP Study of Tamoxifen and Raloxifene (STAR) P-2 trial. JAMA 295(23):2727–2741. https://doi.org/10.1001/jama.295.23.joc60074
Waldman AD, Fritz JM, Lenardo MJ (2020) A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol 20(11):651–668
Wang JJ, Lei KF, Han F (2018) Tumor microenvironment: recent advances in various cancer treatments. Eur Rev Med Pharmacol Sci 22(12):3855–3864. https://doi.org/10.26355/eurrev_201806_15270
Wattenberg LW (1966) Chemoprophylaxis of carcinogenesis: a review. Cancer Res 26:1520–1526
Weinstein IB, Case K (2008) The history of Cancer Research: introducing an AACR Centennial series. Cancer Res 68(17):6861–6862
Yang Y, Arseni D, Zhang W, Huang M, Lövestam S, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Ryskeldi-Falcon B, Scheres SHW, Goedert M (2022) Cryo-EM structures of amyloid-β 42 filaments from human brains. Science (New York, N.Y.) 375(6577):167–172. https://doi.org/10.1126/science.abm7285
Zhang N, Yin Y, Xu SJ, Chen WS (2008) 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules (Basel, Switzerland) 13(8):1551–1569. https://doi.org/10.3390/molecules13081551
Zhao L, Cao YJ (2019) Engineered T Cell Therapy for Cancer in theClinic. Front Immunol 10:2250. https://doi.org/10.3389/fimmu.2019.02250
Zhou Y, Zhou W, Zhou J, Yan J, Xu D, Zheng X, Zong S, Jiang P, Tian S, Han J, Qu D (2022) The Clustered Regularly Interspaced Short Palindromic Repeats-Associated System and Its Relationship With Mobile Genetic Elements in Klebsiella. Front Microbiol 12:790673. https://doi.org/10.3389/fmicb.2021.790673
Zhu YS, Tang K, Lv J (2021) Peptide-drug conjugate-based novel molecular drug delivery system in cancer. Trends Pharmacol Sci 42(10):857–869. https://doi.org/10.1016/j.tips.2021.07.001
Download references
Author information
Authors and affiliations.
Department of Zoology, Panjab University, Chandigarh, India
Indu Sharma, Anuradha Sharma, Reena Tomer & Neha Negi
Department of Biotechnology, Panjab University, Chandigarh, India
Ranbir Chander Sobti
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Indu Sharma .
Editor information
Editors and affiliations.
Dept. Biotechnology, Panjab University, Chandigarh, Chandigarh, India
R. C. Sobti
NCR Biotech Science Cluster, Translational Health Sci & Tech Inst, Faridabad, Haryana, India
Nirmal K. Ganguly
National Chair in Cancer Research, Rajiv Gandhi Center for Biotechnology, trivandrum, Kerala, India
Rakesh Kumar
Section Editor information
Swami Rama Himalayan University, Dehradun, Kerala, India
NCR Biotech Science Cluster, Translational Health Science and Technology Institute, Faridabad, India
Rights and permissions
Reprints and permissions
Copyright information
© 2023 Springer Nature Singapore Pte Ltd.
About this entry
Cite this entry.
Sharma, I., Sharma, A., Tomer, R., Negi, N., Sobti, R.C. (2023). History, Evolution, Milestones in Cancer Research and Treatment. In: Sobti, R.C., Ganguly, N.K., Kumar, R. (eds) Handbook of Oncobiology: From Basic to Clinical Sciences. Springer, Singapore. https://doi.org/10.1007/978-981-99-2196-6_2-1
Download citation
DOI : https://doi.org/10.1007/978-981-99-2196-6_2-1
Received : 08 April 2023
Accepted : 13 April 2023
Published : 15 July 2023
Publisher Name : Springer, Singapore
Print ISBN : 978-981-99-2196-6
Online ISBN : 978-981-99-2196-6
eBook Packages : Springer Reference Biomedicine and Life Sciences Reference Module Biomedical and Life Sciences
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 05 February 2020
The evolutionary history of 2,658 cancers
- Moritz Gerstung 1 , 2 , 3 na1 ,
- Clemency Jolly 4 na1 ,
- Ignaty Leshchiner 5 na1 ,
- Stefan C. Dentro 3 , 4 , 6 na1 ,
- Santiago Gonzalez 1 na1 ,
- Daniel Rosebrock 5 ,
- Thomas J. Mitchell 3 , 7 ,
- Yulia Rubanova 8 , 9 ,
- Pavana Anur 10 ,
- Kaixian Yu 11 ,
- Maxime Tarabichi 3 , 4 ,
- Amit Deshwar 8 , 9 ,
- Jeff Wintersinger 8 , 9 ,
- Kortine Kleinheinz 12 , 13 ,
- Ignacio Vázquez-García 3 , 7 ,
- Kerstin Haase 4 ,
- Lara Jerman 1 , 14 ,
- Subhajit Sengupta 15 ,
- Geoff Macintyre 16 ,
- Salem Malikic 17 , 18 ,
- Nilgun Donmez 17 , 18 ,
- Dimitri G. Livitz 5 ,
- Marek Cmero 19 , 20 ,
- Jonas Demeulemeester 4 , 21 ,
- Steven Schumacher 5 ,
- Yu Fan 11 ,
- Xiaotong Yao 22 , 23 ,
- Juhee Lee 24 ,
- Matthias Schlesner 12 ,
- Paul C. Boutros 8 , 25 , 26 ,
- David D. Bowtell 27 ,
- Hongtu Zhu 11 ,
- Gad Getz 5 , 28 , 29 , 30 ,
- Marcin Imielinski 22 , 23 ,
- Rameen Beroukhim 5 , 31 ,
- S. Cenk Sahinalp 18 , 32 ,
- Yuan Ji 15 , 33 ,
- Martin Peifer 34 ,
- Florian Markowetz 16 ,
- Ville Mustonen 35 ,
- Ke Yuan 16 , 36 ,
- Wenyi Wang 11 ,
- Quaid D. Morris 8 , 9 ,
- PCAWG Evolution & Heterogeneity Working Group ,
- Paul T. Spellman 10 na2 ,
- David C. Wedge 6 , 37 na2 ,
- Peter Van Loo 4 , 21 na2 &
PCAWG Consortium
Nature volume 578 , pages 122–128 ( 2020 ) Cite this article
183k Accesses
532 Citations
836 Altmetric
Metrics details
- Cancer genomics
- Computational biology and bioinformatics
- Molecular evolution
An Author Correction to this article was published on 25 January 2023
This article has been updated
Cancer develops through a process of somatic evolution 1 , 2 . Sequencing data from a single biopsy represent a snapshot of this process that can reveal the timing of specific genomic aberrations and the changing influence of mutational processes 3 . Here, by whole-genome sequencing analysis of 2,658 cancers as part of the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium of the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) 4 , we reconstruct the life history and evolution of mutational processes and driver mutation sequences of 38 types of cancer. Early oncogenesis is characterized by mutations in a constrained set of driver genes, and specific copy number gains, such as trisomy 7 in glioblastoma and isochromosome 17q in medulloblastoma. The mutational spectrum changes significantly throughout tumour evolution in 40% of samples. A nearly fourfold diversification of driver genes and increased genomic instability are features of later stages. Copy number alterations often occur in mitotic crises, and lead to simultaneous gains of chromosomal segments. Timing analyses suggest that driver mutations often precede diagnosis by many years, if not decades. Together, these results determine the evolutionary trajectories of cancer, and highlight opportunities for early cancer detection.
Similar content being viewed by others

Evolutionary signatures of human cancers revealed via genomic analysis of over 35,000 patients
Diletta Fontana, Ilaria Crespiatico, … Daniele Ramazzotti
Pan-cancer analysis of whole genomes
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium

The repertoire of mutational signatures in human cancer
Ludmil B. Alexandrov, Jaegil Kim, … PCAWG Consortium
Similar to the evolution in species, the approximately 10 14 cells in the human body are subject to the forces of mutation and selection 1 . This process of somatic evolution begins in the zygote and only comes to rest at death, as cells are constantly exposed to mutagenic stresses, introducing 1–10 mutations per cell division 2 . These mutagenic forces lead to a gradual accumulation of point mutations throughout life, observed in a range of healthy tissues 5 , 6 , 7 , 8 , 9 , 10 , 11 and cancers 12 . Although these mutations are predominantly selectively neutral passenger mutations, some are proliferatively advantageous driver mutations 13 . The types of mutation in cancer genomes are well studied, but little is known about the times when these lesions arise during somatic evolution and where the boundary between normal evolution and cancer progression should be drawn.
Sequencing of bulk tumour samples enables partial reconstruction of the evolutionary history of individual tumours, based on the catalogue of somatic mutations they have accumulated 3 , 14 , 15 . These inferences include timing of chromosomal gains during early somatic evolution 16 , phylogenetic analysis of late cancer evolution using matched primary and metastatic tumour samples from individual patients 17 , 18 , 19 , 20 , and temporal ordering of driver mutations across many samples 21 , 22 .
The PCAWG Consortium has aggregated whole-genome sequencing data from 2,658 cancers 4 , generated by the ICGC and TCGA, and produced high-accuracy somatic variant calls, driver mutations, and mutational signatures 4 , 23 , 24 (Methods and Supplementary Information ).
Here, we leverage the PCAWG dataset to characterize the evolutionary history of 2,778 cancer samples from 2,658 unique donors across 38 cancer types. We infer timing and patterns of chromosomal evolution and learn typical sequences of mutations across samples of each cancer type. We then define broad periods of tumour evolution and examine how drivers and mutational signatures vary between these epochs. Using clock-like mutational processes, we map mutation timing estimates into approximate real time. Combined, these analyses allow us to sketch out the typical evolutionary trajectories of cancer, and map them in real time relative to the point of diagnosis.
Reconstructing the life history of tumours
The genome of a cancer cell is shaped by the cumulative somatic aberrations that have arisen during its evolutionary past, and part of this history can be reconstructed from whole-genome sequencing data 3 (Fig. 1a ). Initially, each point mutation occurs on a single chromosome in a single cell, which gives rise to a lineage of cells bearing the same mutation. If that chromosomal locus is subsequently duplicated, any point mutation on this allele preceding the gain will subsequently be present on the two resulting allelic copies, unlike mutations succeeding the gain, or mutations on the other allele. As sequencing data enable the measurement of the number of allelic copies, one can define categories of early and late clonal variants, preceding or succeeding copy number gains, as well as unspecified clonal variants, which are common to all cancer cells, but cannot be timed further. Lastly, we identify subclonal mutations, which are present in only a subset of cells and have occurred after the most recent common ancestor (MRCA) of all cancer cells in the tumour sample ( Supplementary Information ).
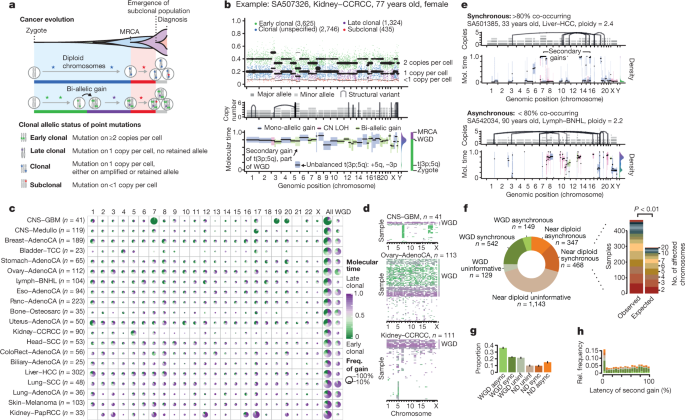
a , Principles of timing mutations and copy number gains based on whole-genome sequencing. The number of sequencing reads reporting point mutations can be used to discriminate variants as early or late clonal (green or purple, respectively) in cases of specific copy number gains, as well as clonal (blue) or subclonal (red) in cases without. b , Annotated point mutations in one sample based on VAF (top), copy number (CN) state and structural variants (middle), and resulting timing estimates (bottom). LOH, loss of heterozygosity. c , Overview of the molecular timing distribution of copy number gains across cancer types. Pie charts depict the distribution of the inferred mutation time for a given copy number gain in a cancer type. Green denotes early clonal gains, with a gradient to purple for late gains. The size of each chart is proportional to the recurrence of this event. Abbreviations for each cancer type are defined in Supplementary Table 1 . d , Heat maps representing molecular timing estimates of gains on different chromosome arms ( x axis) for individual samples ( y axis) for selected tumour types. e , Temporal patterns of two near-diploid cases illustrating synchronous gains (top) and asynchronous gains (bottom). f , Left, distribution of synchronous and asynchronous gain patterns across samples, split by WGD status. Uninformative samples have too few or too small gains for accurate timing. Right, the enrichment of synchronous gains in near-diploid samples is shown by systematic permutation tests. g , Proportion of copy number segments ( n = 90,387) with secondary gains. Error bars denote 95% credible intervals. ND, near diploid. h , Distribution of the relative latency of n = 824 secondary gains with available timing information, scaled to the time after the first gain and aggregated per chromosome.
Source data
The ratio of duplicated to non-duplicated mutations within a gained region can be used to estimate the time point when the gain happened during clonal evolution, referred to here as molecular time, which measures the time of occurrence relative to the total number of (clonal) mutations. For example, there would be few, if any, co-amplified early clonal mutations if the gain had occurred right after fertilization, whereas a gain that happened towards the end of clonal tumour evolution would contain many duplicated mutations 14 (Fig. 1a , Methods).
These analyses are illustrated in Fig. 1b . As expected, the variant allele frequencies (VAFs) of somatic point mutations cluster around the values imposed by the purity of the sample, local copy number configuration and identified subclonal populations. The depicted clear cell renal cell carcinoma has gained chromosome arm 5q at an early molecular time as part of an unbalanced translocation t(3p;5q), which confirms the notion that this lesion often occurs in adolescence in this cancer type 16 . At a later time point, the sample underwent a whole genome duplication (WGD) event, duplicating all alleles, including the derivative chromosome, in a single event, as evidenced by the mutation time estimates of all copy number gains clustering around a single time point, independently of the exact copy number state.
Timing patterns of copy number gains
To systematically examine the mutational timing of chromosomal gains throughout the evolution of tumours in the PCAWG dataset, we applied this analysis to the 2,116 samples with copy number gains suitable for timing ( Supplementary Information ). We find that chromosomal gains occur across a wide range of molecular times (median molecular time 0.60, interquartile range (IQR) 0.10–0.87), with systematic differences between tumour types, whereas within tumour types, different chromosomes typically show similar distributions (Fig. 1c , Extended Data Figs. 1 , 2 , Supplementary Information ). In glioblastoma and medulloblastoma, a substantial fraction of gains occurs early in molecular time. By contrast, in lung cancers, melanomas and papillary kidney cancers, gains arise towards the end of the molecular timescale. Most tumour types, including breast, ovarian and colorectal cancers, show relatively broad periods of chromosomal instability, indicating a very variable timing of gains across samples.
There are, however, certain tumour types with consistently early or late gains of specific chromosomal regions. Most pronounced is glioblastoma, in which 90% of tumours contain single copy gains of chromosome 7, 19 or 20 (Fig. 1c, d ). Notably, these gains are consistently timed within the first 10% of molecular time, which suggests that they arise very early in a patient’s lifetime. In the case of trisomy 7, typically less than 3 out of 600 single nucleotide variants (SNVs) on the whole chromosome precede the gain (Extended Data Fig. 3a, b ). On the basis of a mutation rate of µ = 4.8 × 10 −10 to 3.0 × 10 −9 SNVs per base pair per division 25 , this indicates that the trisomy occurs within the first 6–39 cell divisions, suggesting a possible early developmental origin, in agreement with somatic mosaicisms observed in the healthy brain 26 . Similarly, the duplications leading to isochromosome 17q in medulloblastoma are timed exceptionally early (Extended Data Fig. 3c, d ).
Notably, we observed that gains in the same tumour often appear to occur at a similar molecular time, pointing towards punctuated bursts of copy number gains involving most gained segments (Fig. 1e ). Although this is expected in tumours with WGD (Fig. 1b ), it may seem surprising to observe synchronous gains in near-diploid tumours, particularly as only 6% of co-amplified chromosomal segments were linked by a direct inter-chromosomal structural variant. Still, synchronous gains are frequent, occurring in 57% (468 out of 815) of informative near-diploid tumours, 61% more frequently than expected by chance ( P < 0.01, permutation test; Fig. 1f ). Because most arm-level gains increment the allele-specific copy number by 1 (80–90%; Fig. 1g ), it seems that these gains arise through mis-segregation of single copies during anaphase. This notion is further supported by the observation that in about 85% of segments with two gains of the same allele, the second gain appears with noticeable latency after the first (Fig. 1h ). Therefore, the extensive chromosome-scale copy number aberrations observed in many cancer genomes are seemingly caused by a limited number of events—possibly by merotelic attachments of chromosomes to multipolar mitotic spindles 27 , or as a consequence of negative selection of individual aneuploidies 28 —offering an explanation for observations of punctuated evolution in breast and colorectal cancer 29 , 30 .
Timing of point mutations in driver genes
As outlined above, point mutations (SNVs and insertions and deletions (indels)) can be qualitatively assigned to different epochs, allowing the timing of driver mutations. Out of the 47 million point mutations in 2,583 unique samples, 22% were early clonal, 7% late clonal, 53% unspecified clonal and 17% subclonal (Fig. 2a ). Among a panel of 453 cancer driver genes, 5,913 oncogenic point mutations were identified 4 , of which 29% were early clonal, 5% late clonal, 56% unspecified clonal and 8% subclonal. It thus emerges that common drivers are enriched in the early clonal and unspecified clonal categories and depleted in the late clonal and subclonal ones, indicating a preferential early timing (Fig. 2b ). For example, driver mutations in TP53 and KRAS are 12 and 8 times enriched in early clonal stages, respectively. For TP53 , this trend is independent of tumour type (Fig. 2c ). Mutations in PIK3CA are two times more frequently clonal than expected, and non-coding changes near the TERT gene are three times more frequently early clonal.
a , Top, distribution of point mutations over different mutation periods in n = 2,778 samples. Middle, timing distribution of driver mutations in the 50 most recurrent lesions across n = 2,583 white listed samples from unique donors. Bottom, distribution of driver mutations across cancer types; colour as defined in the inset. b , Relative timing of the 50 most recurrent driver lesions, calculated as the odds ratio of early versus late clonal driver mutations versus background, or clonal versus subclonal. Error bars denote 95% confidence intervals derived from bootstrap resampling. Odds ratios overlapping 1 in less than 5% of bootstrap samples are considered significant (coloured). The underlying number of samples with a given mutation is shown in a . c , Relative timing of TP53 mutations across cancer types, as in b . The number of samples is defined in the x -axis labels. d , Estimated number of unique lesions (genes) contributing 50% of all driver mutations in different timing epochs across n = 2,583 unique samples, containing n = 5,756 driver mutations with available timing information. Error bars denote the range between 0 and 1 pseudocounts; bars denote the average of the two values. NA, not applicable; NS, not significant.
Aggregating the clonal status of all driver point mutations over time reveals an increased diversity of driver genes mutated at later stages of tumour development: 50% of all early clonal driver mutations occur in just 9 genes, whereas 50% of late and subclonal mutations occur in approximately 35 different genes each, a nearly fourfold increase (Fig. 2d ). Consistent with previous studies of individual tumour types 31 , 32 , 33 , 34 , these results suggest that, in general, the very early events in cancer evolution occur in a constrained set of common drivers, and a more diverse array of drivers is involved in late tumour development.
Relative timing of somatic driver events
Although timing estimates of individual events reflect evolutionary periods that differ from one sample to another, they define in part the order in which driver mutations and copy number alterations have occurred in each sample (Fig. 3a–d ). As confirmed by simulations, aggregating these orderings across samples defines a probabilistic ranking of lesions (Fig. 3a ), recapitulating whether each mutation occurs preferentially early or late during tumour evolution (Extended Data Figs. 4 , 5 , Supplementary Information ).
a , Schematic representation of the ordering process. b – d , Examples of individual patient trajectories (partial ordering relationships), the constituent data for the ordering model process. e – g , Preferential ordering diagrams for colorectal adenocarcinoma (ColoRect–AdenoCA) ( e ), pancreatic neuroendocrine cancer (Panc–Endocrine) ( f ) and glioblastoma (CNS–GBM) ( g ). Probability distributions show the uncertainty of timing for specific events in the cohort. Events with odds above 10 (either earlier or later) are highlighted. The prevalence of the event type in the cohort is displayed as a bar plot on the right.
In colorectal adenocarcinoma, for example, we find APC mutations to have the highest odds of occurring early, followed by KRAS , loss of 17p and TP53 , and SMAD4 (Fig. 3b , e). Whole-genome duplications occur after tumours have accumulated several driver mutations, and many chromosomal gains and losses are typically late. These results are in agreement with the classical APC-KRAS-TP53 progression model of Fearon and Vogelstein 35 , but add considerable detail.
In many cancer types, the sequence of events during cancer progression has not previously been determined in detail. For example, in pancreatic neuroendocrine cancers, we find that many chromosomal losses, including those of chromosomes 2, 6, 11 and 16, are among the earliest events, followed by driver mutations in MEN1 and DAXX (Fig. 3c, f ). WGD events occur later, after many of these tumours have reached a pseudo-haploid state due to widespread chromosomal losses. In glioblastoma, we find that the loss of chromosome 10, and driver mutations in TP53 and EGFR are very early, often preceding early gains of chromosomes 7, 19 and 20 (Fig. 3d, g ). Mutations in the TERT promoter tend to occur at early to intermediate time points, whereas other driver mutations and copy number changes tend to be later events.
Across cancer types, we typically find TP53 mutations among the earliest events, as well as losses of chromosome 17 ( Supplementary Information ). WGD events usually have an intermediate ranking, and most copy number changes occur later. Losses typically precede gains, and consistent with the results above, common drivers typically occur before rare drivers.
Timing of mutational signatures
The cancer genome is shaped by various mutational processes over its lifetime, stemming from exogenous and cell-intrinsic DNA damage, and error-prone DNA replication, leaving behind characteristic mutational spectra, termed mutational signatures 24 , 36 . Stratifying mutations by their clonal allelic status, we find evidence for a changing mutational spectrum between early and late clonal time points in 29% (530 out of 1,852) of informative samples ( P < 0.05, Bonferroni-adjusted likelihood-ratio test), typically changing the spectrum by 19% (median absolute difference; range 4–66%) (Fig. 4a, b , Extended Data Fig. 6 ). Similarly, 30% of informative samples (729 out of 2,387) displayed changes of their mutation spectrum between the clonal and subclonal state, with median difference of 21% (range 3–72%). Combined, the mutation spectrum changes throughout tumour evolution in 40% of samples (1,069 out of 2,688).
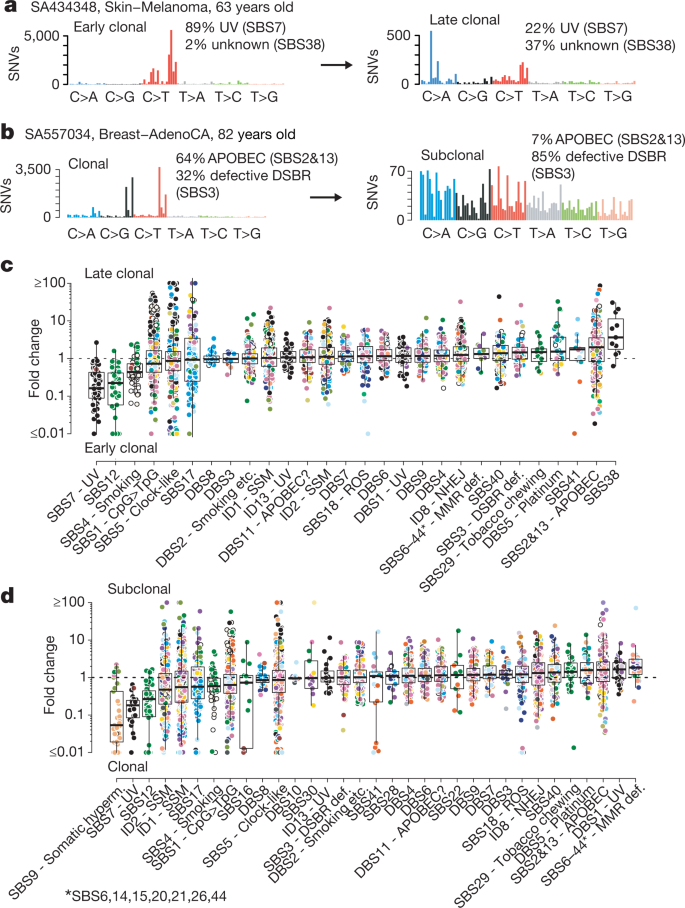
a , Example of tumours with substantial changes between mutation spectra of early (left) and late (right) clonal time points. The attribution of mutations to the most characteristic signatures are shown. b , Example of clonal-to-subclonal mutation spectrum change. c , Fold changes between relative proportions of early and late clonal mutations attributed to individual mutational signatures. Points are coloured by tissue type. Data are shown for samples ( n = 530) with measurable changes in their overall mutation spectra and restricted to signatures active in at least 10 samples. Box plots demarcate the first and third quartiles of the distribution, with the median shown in the centre and whiskers covering data within 1.5× the IQR from the box. d , Fold changes between clonal and subclonal periods in samples ( n = 729) with measurable changes in their mutation spectra, analogous to c .
To quantify whether the observed temporal changes can be attributed to known and suspected mutational processes, we decomposed the mutational spectra at each time point into a catalogue of 57 mutational signatures, including double base substitution and indel signatures 24 (Methods).
In general, these mutational signatures display a predominantly undirected temporal variability over several orders of magnitude (Fig. 4c, d , Extended Data Fig. 7 ). In addition, several signatures demonstrate distinct temporal trends. As one may expect, signatures of exogenous mutagens are predominantly active in the early clonal stages of tumorigenesis. These include tobacco smoking in lung adenocarcinoma (signature SBS4, median fold change 0.43, IQR 0.31–0.72), consistent with previous reports 37 , 38 , and ultraviolet light exposure in melanoma (SBS7; median fold change 0.16, IQR 0.09–0.43). Another strong decrease over time is found for a signature of unknown aetiology, SBS12, which acts mostly in liver cancers (median fold change 0.22, IQR 0.06–0.41). In chronic lymphoid leukaemia, there was a 20-fold relative decrease in mutations associated with somatic hypermutation (SBS9; median fold change 0.05, IQR 0.02–0.43) from clonal to subclonal stages.
Some mutational processes tend to increase throughout cancer evolution. For example, we see that APOBEC mutagenesis (SBS2 and SBS13) increases in many cancer types from the early to late clonal stages (median fold change 2.0, IQR 0.8–3.6), as does a newly described signature SBS38 (median fold 3.6, IQR 1.8–11). Signatures of defective mismatch repair (SBS6, 14, 15, 20, 21, 26 and 44) increase from clonal to subclonal stages (median fold 1.8, IQR 1.2–3.0).
Chronological time estimates
The molecular timing data presented above do not measure the occurrence of events in chronological time. If the rate at which mutations are acquired per year in each sample was constant, the chronological time would simply be the product of the estimated molecular timing and age at diagnosis. However, this relation will be nonlinear if the mutation rate changes over time, and is inflated by acquired mutational processes, as suggested by the analysis in the previous section. Some of these issues can be mitigated by counting only mutations contributed by endogenous and less variable mutational processes, such as CpG-to-TpG mutations (hereafter CpG>TpG) caused by spontaneous deamination of 5-methyl-cytosine to thymine at CpG dinucleotides, which have been proposed as a molecular clock 12 . Our supplementary analysis suggests that, although the baseline CpG>TpG mutation rate in cancers is very close to that in normal cells, there appears to be a moderate increase (1–10 times, adding between 20 and 40% of mutations) in cancers (Extended Data Fig. 8 ). As this shifts chronological timing estimates, we model different scenarios of the evolution of the CpG>TpG mutation rate (Fig. 5a ).
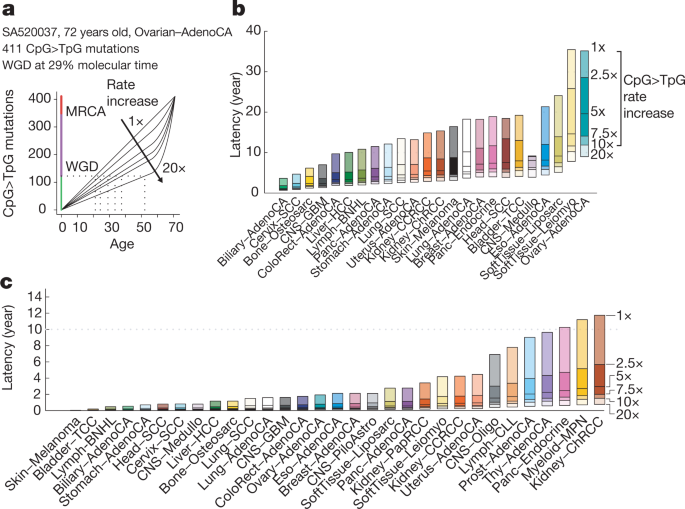
a , Mapping of molecular timing estimates to chronological time under different scenarios of increases in the CpG>TpG mutation rate. A greater increase before diagnosis indicates an inflation of the mutation timescale. b , Median latency between WGDs and the last detectable subclone before diagnosis under different scenarios of CpG>TpG mutation rate increases for n = 569 non-hypermutant cancers with at least 100 informative SNVs, low tumour in normal contamination and at least five samples per tumour histology. c , Median latency between the MRCA and the last detectable subclone before diagnosis for different CpG>TpG mutation rate changes in n = 1,921 non-hypermutant samples with low tumour in normal contamination and at least 5 cases per cancer type.
Applying this logic to time WGDs, which yield sufficient numbers of CpG>TpG mutations, demonstrates that they occur several years and possibly even a decade or more before diagnosis in some cancer types, under a range of scenarios of mutation rate increase (Fig. 5b , Extended Data Fig. 9 ). A notable example is ovarian adenocarcinoma, which appears to have a median latency of more than 10 years. This holds true even under a scenario of a CpG>TpG rate increase of 20-fold, which would be far beyond the 7.5-fold rate increase observed in matched primary and relapse samples 39 (Extended Data Fig. 8f ). Notably, these results suggest WGD may occur throughout the entire female reproductive life (Extended Data Fig. 9b ). The latency between the MRCA and the last detectable subclone is shorter, typically several months to years (Fig. 5c ).
These timescales of cancer evolution are further supported by the fact that progression of most known precancerous lesions to carcinomas usually spans many years, if not decades 40 , 41 , 42 , 43 , 44 , 45 . Our data corroborate these timescales and extend them to cancer types without detectable premalignant conditions, raising the hope that these tumours could also be detected in less malignant stages.
To our knowledge, our study presents the first large-scale genome-wide reconstruction of the evolutionary history of cancers, reconstructing both early (pre-cancer) and later stages of 38 cancer types. This is facilitated by the timing of copy number gains relative to all other events in the genome, through multiplicity and clonal status of co-amplified point mutations. However, several limitations exist ( Supplementary Information ). Perhaps most importantly, molecular timing is based on point mutations and is therefore subject to changes in mutation rate. Notably, healthy tissues acquire point mutations at rates not too dissimilar from those seen in cancers, particularly when considering only endogenous mutational processes, and furthermore, some tissues are riddled with microscopic clonal expansions of driver gene mutations 5 , 6 , 7 , 8 , 9 , 11 . This is direct evidence that the life history of almost every cell in the human body, including those that develop into cancer, is driven by somatic evolution.
Together, the data presented here enable us to draw approximate timelines summarizing the typical evolutionary history of each cancer type (Fig. 6 , Supplementary Information for all other cancer types). These make use of the qualitative timing of point mutations and copy number alterations, as well as signature activities, which can be interleaved with the chronological estimates of WGD and the appearance of the MRCA.
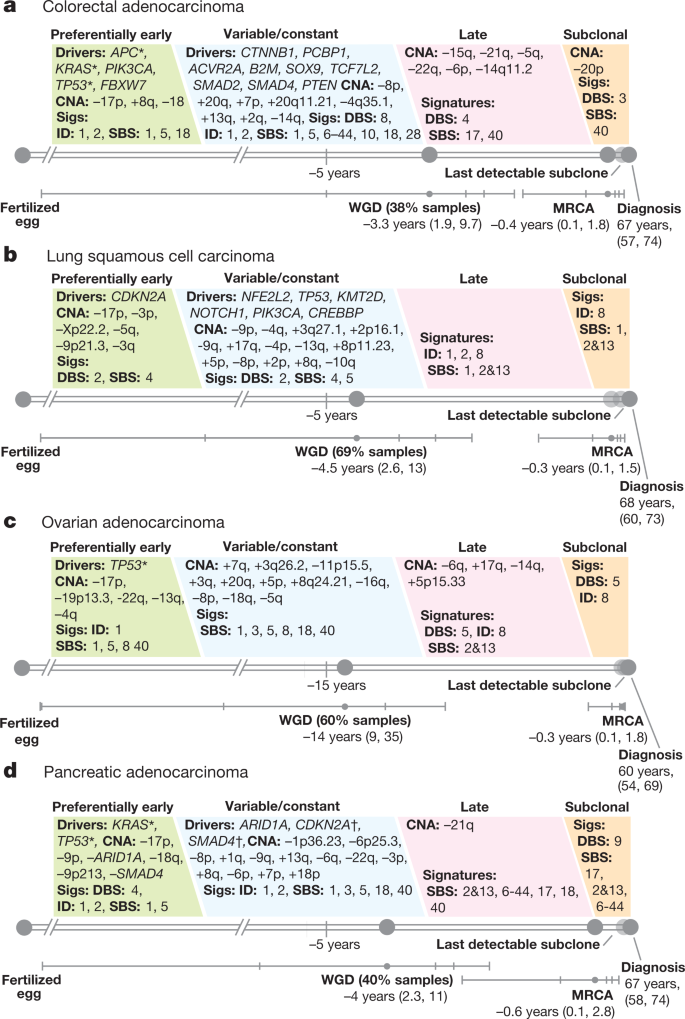
a – d , Timelines representing the length of time, in years, between the fertilized egg and the median age of diagnosis for colorectal adenocarcinoma ( a ), squamous cell lung cancer ( b ), ovarian adenocarcinoma ( c ) and pancreatic adenocarcinoma ( d ). Real-time estimates for major events, such as WGD and the emergence of the MRCA, are used to define early, variable, late and subclonal stages of tumour evolution approximately in chronological time. The range of chronological time estimates according to varying clock mutation acceleration rates is shown as well, with tick marks corresponding to 1×, 2.5×, 5×, 7.5×, 10× and 20×. Driver mutations and copy number alterations (CNA) are shown in each stage according to their preferential timing, as defined by relative ordering. Mutational signatures (Sigs) that, on average, change over the course of tumour evolution, or are substantially active but not changing, are shown in the epoch in which their activity is greatest. DBS, double base substitution; SBS, single base substitutions. Where applicable, lesions with a known timing from the literature are annotated; dagger symbols denotes events that were found to have a different timing; asterisk symbol denotes events that agree with our timing.
It is remarkable that the evolution of practically all cancers displays some level of order, which agrees very well with, and adds much detail to, established models of cancer progression 35 , 46 . For example, TP53 with accompanying 17p deletion is one of the most frequent initiating mutations in a variety of cancers, including ovarian cancer, in which it is the hallmark of its precancerous precursor lesions 47 . Furthermore, the list of typically early drivers includes most other highly recurrent cancer genes, such as KRAS , TERT and CDKN2A , indicating a preferred role in early and possibly even pre-cancer evolution. This initially constrained set of genes broadens at later stages of cancer development, suggesting an epistatic fitness landscape canalizing the first steps of cancer evolution. Over time, as tumours evolve, they follow increasingly diverse paths driven by individually rare driver mutations, and by copy number alternations. However, none of these trends is absolute, and the evolutionary paths of individual tumours are highly variable, showing that cancer evolution follows trends, but is far from deterministic.
Our study sheds light on the typical timescales of in vivo tumour development, with initial driver events seemingly occurring up to decades before diagnosis, demonstrating how cancer genomes are shaped by a lifelong process of somatic evolution, with fluid boundaries between normal ageing processes 5 , 6 , 7 , 8 , 9 , 10 , 11 and cancer evolution. Nevertheless, the presence of genetic aberrations with such long latency raises hopes that aberrant clones could be detected early, before reaching their full malignant potential.
The PCAWG series consists of 2,778 tumour samples (2,703 white listed, 75 grey listed) from 2,658 donors. All samples in this dataset underwent whole-genome sequencing (minimum average coverage 30× in the tumour, 25× in the matched normal samples), and were processed with a set of project-specific pipelines for alignment, variant calling, and quality control 4 . Copy number calls were established by combining the output of six individual callers into a consensus using a multi-tier approach, resulting in a copy number profile, a purity and ploidy value and whether the tumour has undergone a WGD ( Supplementary Information ). Consensus subclonal architectures have been obtained by integrating the output of 11 subclonal reconstruction callers, after which all SNVs, indels and structural variants are assigned to a mutation cluster using the MutationTimer.R approach ( Supplementary Information ). Driver calls have been defined by the PCAWG Driver Working Group 4 , and mutational signatures are defined by the PCAWG Signatures Working Group 24 . A more detailed description can be found in Supplementary Information, section 1 .
Data accrual was based on sequencing experiments performed by individual member groups of the ICGC and TCGA, as described in an associated study 4 . As this is a meta-analysis of existing data, power calculations were not performed and the investigators were not blinded to cancer diagnoses.
Timing of gains
We used three related approaches to calculate the timing of copy number gains (see Supplementary Information, section 2 ). In brief, the common feature is that the expected VAF of a mutation ( E ) is related to the underlying number of alleles carrying a mutation according to the formula: E [ X ] = nmfρ /[ N (1 − ρ ) + Cρ ], in which X is the number of reads, n denotes the coverage of the locus, the mutation copy number m is the number of alleles carrying the mutation (which is usually inferred), f is the frequency of the clone carrying the given mutation ( f = 1 for clonal mutations). N is the normal copy number (2 on autosomes, 1 or 2 for chromosome X and 0 or 1 for chromosome Y), C is the total copy number of the tumour, and ρ is the purity of the sample.
The number of mutations n m at each allelic copy number m then informs about the time when the gain has occurred. The basic formulae for timing each gain are, depending on the copy number configuration:
in which 2 + 1 refers to major and minor copy number of 2 and 1, respectively. Methods differ slightly in how the number of mutations present on each allele are calculated and how uncertainty is handled ( Supplementary Information ).
Timing of mutations
The mutation copy number m and the clonal frequency f is calculated according to the principles indicated above. Details can be found in Supplementary Information, section 2 . Mutations with f = 1 are denoted as ‘clonal’, and mutations with f < 1 as ‘subclonal’. Mutations with f = 1 and m > 1 are denoted as ‘early clonal’ (co-amplified). In cases with f = 1, m = 1 and C > 2, mutations were annotated as ‘late clonal’, if the minor copy number was 0, otherwise ‘clonal’ (unspecified).
Timing of driver mutations
A catalogue of driver point mutations (SNVs and indels) was provided by the PCAWG Drivers and Functional Interpretation Group 4 . The timing category was calculated as above. From the four timing categories, the odds ratios of early/late clonal and clonal (early, late or unspecified clonal)/subclonal were calculated for driver mutations against the distribution of all other mutations present in fragments with the same copy number composition in the samples with each particular driver. The background distribution of these odds ratios was assessed with 1,000 bootstraps ( Supplementary Information, section 4.1 ).
Integrative timing
For each pair of driver point mutations and recurrent copy number alterations, an ordering was established (earlier, later or unspecified). The information underlying this decision was derived from the timing of each driver point mutation, as well as from the timing status of clonal and subclonal copy number segments. These tables were aggregated across all samples and a sports statistics model was employed to calculate the overall ranking of driver mutations. A full description is given in Supplementary Information, section 4.2 .
Mutational trinucleotide substitution signatures, as defined by the PCAWG Mutational Signatures Working Group 24 , were fit to samples with observed signature activity, after splitting point mutations into either of the four epochs. A likelihood ratio test based on the multinomial distribution was used to test for differences in the mutation spectra between time points. Time-resolved exposures were calculated using non-negative linear least squares. Full details are given in Supplementary Information, section 5 .
Real-time estimation of WGD and MRCA
CpG>TpG mutations were counted in an NpCpG context, except for skin–melanoma, in which CpCpG and TpCpG were excluded owing to the overlapping UV mutation spectrum. For visual comparison, the number of mutations was scaled to the effective genome size, defined as the 1/mean( m i / C i ), in which m i is the estimated number of allelic copies of each mutation, and C i is the total copy number at that locus, thereby scaling to the final copy number and the time of change.
A hierarchical Bayesian linear regression was fit to relate the age at diagnosis to the scaled number of mutations, ensuring positive slope and intercept through a shared gamma distribution across cancer types.
For tumours with several time points, the set of mutations shared between diagnosis and relapse ( n D ) and those specific to the relapse ( n R ) was calculated. The rate acceleration was calculated as: a = n R / n D × t D / t R . This analysis was performed separately for all substitutions and for CpG>TpG mutations.
On the basis of these analyses, a typical increase of 5× for most cancer types was chosen, with a lower value of 2.5× for brain cancers and a value of 7.5× for ovarian cancer.
The correction for transforming an estimate of a copy number gain in mutation time into chronological time depends not only on the rate acceleration, but also on the time at which this acceleration occurred. As this is generally unknown, we performed Monte Carlo simulations of rate accelerations spanning an interval of 15 years before diagnosis, corresponding roughly to 25% of time for a diagnosis at 60 years of age, noting that a 5× rate increase over this duration yields an offset of about 33% of mutations, compatible with our data. Subclonal mutations were assumed to occur at full acceleration. The proportion of subclonal mutations was divided by the number of identified subclones, thus conservatively assuming branching evolution. Full details are given in Supplementary Information, section 6 .
Cancer timelines
The results from each of the different timing analyses are combined in timelines of cancer evolution for each tumour type (Fig. 6 and Supplementary Information ). Each timeline begins at the fertilized egg, and spans up to the median age of diagnosis within each cohort. Real-time estimates for WGD and the MRCA act as anchor points, allowing us to roughly map the four broadly defined time periods (early clonal, intermediate, late clonal and subclonal) to chronological time during a patient’s lifespan. Specific driver mutations or copy number alterations can be placed within each of these time frames based on their ordering from the league model analysis. Signatures are shown if they typically change over time (95% confidence intervals of mean change not overlapping 0), and if they are strongly active (contributing at least 10% mutations to one time point). Signatures are shown on the timeline in the epoch of their greatest activity. Where an event found in our study has a known timing in the literature, the agreement is annotated on the timeline; with an asterisk denoting an agreed timing, and dagger symbol denoting a timing that is different to our results. Full details are given in Supplementary Information, section 7 .
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Somatic and germline variant calls, mutational signatures, subclonal reconstructions, transcript abundance, splice calls and other core data generated by the ICGC/TCGA PCAWG Consortium are described elsewhere 4 and available for download at https://dcc.icgc.org/releases/PCAWG . Further information on accessing the data, including raw read files, can be found at https://docs.icgc.org/pcawg/data/ . In accordance with the data access policies of the ICGC and TCGA projects, most molecular, clinical and specimen data are in an open tier that does not require access approval. To access information that could potentially identify participants, such as germline alleles and underlying sequencing data, researchers will need to apply to the TCGA Data Access Committee (DAC) via dbGaP ( https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login ) for access to the TCGA portion of the dataset, and to the ICGC Data Access Compliance Office (DACO; http://icgc.org/daco ) for the ICGC portion. In addition, to access somatic SNVs derived from TCGA donors, researchers will also need to obtain dbGaP authorization. Datasets used and results presented in this study, including timing estimates for copy number gains, chronological estimates of WGD and MRCA, as well as mutation signature changes, are described in Supplementary Note 3 and are available at https://dcc.icgc.org/releases/PCAWG/evolution-heterogeneity .
Code availability
The core computational pipelines used by the PCAWG Consortium for alignment, quality control and variant calling are available to the public at https://dockstore.org/search?search=pcawg under the GNU General Public License v3.0, which allows for reuse and distribution. Analysis code presented in this study is available through the GitHub repository https://github.com/PCAWG-11/Evolution . This archive contains relevant software and analysis workflows as submodules, which include code for timing copy number gains, point mutations and mutation signatures, real-time timing and evolutionary league model analysis, as well as scripts to generate the figures presented: CancerTiming (v.3.1.8), MutationTimeR (v.0.1), PhylogicNDT (v.1.1) and a series of custom scripts (v. 1.0), with detailed versions of other packages used.
Change history
25 january 2023.
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-05601-4
Cairns, J. Mutation selection and the natural history of cancer. Nature 255 , 197–200 (1975).
Article ADS CAS Google Scholar
Martincorena, I. & Campbell, P. J. Somatic mutation in cancer and normal cells. Science 349 , 1483–1489 (2015).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149 , 994–1007 (2012).
Article CAS Google Scholar
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature https://doi.org/10.1038/s41586-020-1969-6 (2020).
Moore, L. et al. The mutational landscape of normal human endometrial epithelium. Preprint at bioRxiv https://doi.org/10.1101/505685 (2018).
Lee-Six, H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574 , 532–537 (2019).
Lee-Six, H. et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561 , 473–478 (2018).
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362 , 911–917 (2018).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348 , 880–886 (2015).
Welch, J. S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150 , 264–278 (2012).
Yokoyama, A. et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565 , 312–317 (2019).
Alexandrov, L. B. et al. Clock-like mutational processes in human somatic cells. Nat. Genet . 47 , 1402–1407 (2015).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194 , 23–28 (1976).
Durinck, S. et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov . 1 , 137–143 (2011).
Jolly, C. & Van Loo, P. Timing somatic events in the evolution of cancer. Genome Biol . 19 , 95 (2018).
Article Google Scholar
Mitchell, T. J. et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx Renal. Cell 173 , 611–623 (2018).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med . 366 , 883–892 (2012).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520 , 353–357 (2015).
Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med . 21 , 751–759 (2015).
Brastianos, P. K. et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov . 5 , 1164–1177 (2015).
Papaemmanuil, E. et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122 , 3616–3627 (2013).
Landau, D. A. et al. Mutations driving CLL and their evolution in progression and relapse. Nature 526 , 525–530 (2015).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature https://doi.org/10.1038/s41586-020-1965-x (2020).
Alexandrov, L. B. The repertoire of mutational signatures in human cancer. Nature https://doi.org/10.1038/s41586-020-1943-3 (2020).
Keogh, M. J. et al. High prevalence of focal and multi-focal somatic genetic variants in the human brain. Nat. Commun . 9 , 4257 (2018).
Article ADS Google Scholar
Heim, S. et al. Trisomy 7 and sex chromosome loss in human brain tissue. Cytogenet. Cell Genet . 52 , 136–138 (1989).
Ganem, N. J., Godinho, S. A. & Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 460 , 278–282 (2009).
Sheltzer, J. M. et al. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31 , 240–255 (2017).
Gao, R. et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet . 48 , 1119–1130 (2016).
Cross, W. et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2 , 1661–1672 (2018).
Gerlinger, M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet . 46 , 225–233 (2014).
Gibson, W. J. et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat. Genet . 48 , 848–855 (2016).
Yates, L. R. et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32 , 169–184 (2017).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med . 376 , 2109–2121 (2017).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61 , 759–767 (1990).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500 , 415–421 (2013).
McGranahan, N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med . 7 , 283ra54 (2015).
Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol . 17 , 31 (2016).
Patch, A.-M. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521 , 489–494 (2015).
Bostwick, D. G. & Qian, J. High-grade prostatic intraepithelial neoplasia. Mod. Pathol . 17 , 360–379 (2004).
Brenner, H. et al. Risk of progression of advanced adenomas to colorectal cancer by age and sex: estimates based on 840,149 screening colonoscopies. Gut 56 , 1585–1589 (2007).
Gazdar, A. F. & Brambilla, E. Preneoplasia of lung cancer. Cancer Biomark . 9 , 385–396 (2010).
Sanders, M. E., Schuyler, P. A., Dupont, W. D. & Page, D. L. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer 103 , 2481–2484 (2005).
Schlecht, N. F. et al. Human papillomavirus infection and time to progression and regression of cervical intraepithelial neoplasia. J. Natl. Cancer Inst . 95 , 1336–1343 (2003).
Whitson, M. J. & Falk, G. W. Predictors of progression to high-grade dysplasia or adenocarcinoma in Barrett’s esophagus. Gastroenterol. Clin. North Am . 44 , 299–315 (2015).
Bardeesy, N. & DePinho, R. A. Pancreatic cancer biology and genetics. Nat. Rev. Cancer 2 , 897–909 (2002).
Folkins, A. K. et al. A candidate precursor to pelvic serous cancer (p53 signature) and its prevalence in ovaries and fallopian tubes from women with BRCA mutations. Gynecol. Oncol . 109 , 168–173 (2008).
Download references
Acknowledgements
We thank H. Lee-Six and L. Moore for sharing data on mutation burden in normal tissues. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001202), the UK Medical Research Council (FC001202) and the Wellcome Trust (FC001202). This project was enabled through the Crick Scientific Computing STP and through access to the MRC eMedLab Medical Bioinformatics infrastructure, supported by the Medical Research Council (grant number MR/L016311/1). M.T. and J.D. are postdoctoral fellows supported by the European Union’s Horizon 2020 research and innovation program (Marie Skłodowska-Curie grant agreement number 747852-SIOMICS and 703594-DECODE). J.D. is a postdoctoral fellow of the FWO. F.M., G.M. and K. Yuan acknowledge the support of the University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited. G.M., K. Yuan and F.M. were funded by CRUK core grants C14303/A17197 and A19274. S. Sengupta and Y.J. are supported by NIH R01 CA132897. S.M. is supported by the Vanier Canada Graduate Scholarship. S.C.S. is supported by the NSERC Discovery Frontiers Project, “The Cancer Genome Collaboratory” and NIH Grant GM108308. H.Z. is supported by grant NIMH086633 and an endowed Bao-Shan Jing Professorship in Diagnostic Imaging. W.W. is supported by the US National Cancer Institute (1R01 CA183793 and P30 CA016672). P.T.S. was supported by U24CA210957 and 1U24CA143799. D.C.W. is funded by the Li Ka Shing foundation. P.V.L. is a Winton Group Leader in recognition of the Winton Charitable Foundation’s support towards the establishment of The Francis Crick Institute. We acknowledge the contributions of the many clinical networks across ICGC and TCGA who provided samples and data to the PCAWG Consortium, and the contributions of the Technical Working Group and the Germline Working Group of the PCAWG Consortium for collation, realignment and harmonized variant calling of the cancer genomes used in this study. We thank the patients and their families for their participation in the individual ICGC and TCGA projects.
Author information
These authors contributed equally: Moritz Gerstung, Clemency Jolly, Ignaty Leshchiner, Stefan C. Dentro, Santiago Gonzalez
These authors jointly supervised this work: Paul T. Spellman, David C. Wedge, Peter Van Loo
A list of members and their affiliations appears at the end of the paper
A list of members and their affiliations appears online
Authors and Affiliations
European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Cambridge, UK
Moritz Gerstung, Santiago Gonzalez, Lara Jerman, Moritz Gerstung, Santiago Gonzalez & Lara Jerman
European Molecular Biology Laboratory, Genome Biology Unit, Heidelberg, Germany
Moritz Gerstung & Moritz Gerstung
Wellcome Sanger Institute, Cambridge, UK
Moritz Gerstung, Stefan C. Dentro, Thomas J. Mitchell, Maxime Tarabichi, Ignacio Vázquez-García, Stefan C. Dentro, Moritz Gerstung, Maxime Tarabichi, David J. Adams, Peter J. Campbell, Kevin J. Dawson, Henry Lee-Six, Inigo Martincorena, Thomas J. Mitchell & Ignacio Vázquez-García
The Francis Crick Institute, London, UK
Clemency Jolly, Stefan C. Dentro, Maxime Tarabichi, Kerstin Haase, Jonas Demeulemeester, Stefan C. Dentro, Clemency Jolly, Kerstin Haase, Maxime Tarabichi, Jonas Demeulemeester, Matthew Fittall, Peter Van Loo & Peter Van Loo
Broad Institute of MIT and Harvard, Cambridge, MA, USA
Ignaty Leshchiner, Daniel Rosebrock, Dimitri G. Livitz, Steven Schumacher, Gad Getz, Rameen Beroukhim, Ignaty Leshchiner, Rameen Beroukhim, Gad Getz, Gavin Ha, Dimitri G. Livitz, Daniel Rosebrock, Steven Schumacher & Oliver Spiro
Big Data Institute, University of Oxford, Oxford, UK
Stefan C. Dentro, Stefan C. Dentro, David C. Wedge & David C. Wedge
University of Cambridge, Cambridge, UK
Thomas J. Mitchell, Ignacio Vázquez-García, Thomas J. Mitchell & Ignacio Vázquez-García
University of Toronto, Toronto, Ontario, Canada
Yulia Rubanova, Amit Deshwar, Jeff Wintersinger, Paul C. Boutros, Quaid D. Morris, Jeff Wintersinger, Amit G. Deshwar, Yulia Rubanova, Paul C. Boutros, Ruian Shi, Shankar Vembu & Quaid D. Morris
Vector Institute, Toronto, Ontario, Canada
Yulia Rubanova, Amit Deshwar, Jeff Wintersinger, Quaid D. Morris, Jeff Wintersinger, Amit G. Deshwar, Yulia Rubanova & Quaid D. Morris
Molecular and Medical Genetics, Oregon Health & Science University, Portland, OR, USA
Pavana Anur, Pavana Anur, Myron Peto, Paul T. Spellman & Paul T. Spellman
The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Kaixian Yu, Yu Fan, Hongtu Zhu, Wenyi Wang, Kaixian Yu, Shaolong Cao, Yu Fan, Seung Jun Shin, Hongtu Zhu & Wenyi Wang
German Cancer Research Center (DKFZ), Heidelberg, Germany
Kortine Kleinheinz, Matthias Schlesner, Roland Eils, Kortine Kleinheinz & Matthias Schlesner
Heidelberg University, Heidelberg, Germany
Kortine Kleinheinz, Roland Eils & Kortine Kleinheinz
University of Ljubljana, Ljubljana, Slovenia
Lara Jerman & Lara Jerman
NorthShore University HealthSystem, Evanston, IL, USA
Subhajit Sengupta, Yuan Ji, Yuan Ji & Subhajit Sengupta
Cancer Research UK Cambridge Institute, University of Cambridge, Cambridge, UK
Geoff Macintyre, Florian Markowetz, Ke Yuan, Geoff Macintyre, Ruben M. Drews, Florian Markowetz & Ke Yuan
Simon Fraser University, Burnaby, British Columbia, Canada
Salem Malikic, Nilgun Donmez, Nilgun Donmez & Salem Malikic
Vancouver Prostate Centre, Vancouver, British Columbia, Canada
Salem Malikic, Nilgun Donmez, S. Cenk Sahinalp, Nilgun Donmez, Salem Malikic & S. Cenk Sahinalp
University of Melbourne, Melbourne, Victoria, Australia
Marek Cmero, Elizabeth L. Christie, Marek Cmero & Dale W. Garsed
Walter and Eliza Hall Institute, Melbourne, Victoria, Australia
Marek Cmero & Marek Cmero
University of Leuven, Leuven, Belgium
Jonas Demeulemeester, Jonas Demeulemeester, Peter Van Loo & Peter Van Loo
Weill Cornell Medicine, New York, NY, USA
Xiaotong Yao, Marcin Imielinski, Marcin Imielinski & Xiaotong Yao
New York Genome Center, New York, NY, USA
University of California Santa Cruz, Santa Cruz, CA, USA
Juhee Lee & Juhee Lee
Ontario Institute for Cancer Research, Toronto, Ontario, Canada
Paul C. Boutros, Paul C. Boutros, Adriana Salcedo & Lincoln D. Stein
University of California, Los Angeles, CA, USA
Paul C. Boutros & Paul C. Boutros
Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia
David D. Bowtell, David D. Bowtell, Elizabeth L. Christie & Dale W. Garsed
Center for Cancer Research, Massachusetts General Hospital, Charlestown, MA, USA
Gad Getz & Gad Getz
Department of Pathology, Massachusetts General Hospital, Boston, MA, USA
Harvard Medical School, Boston, MA, USA
Dana-Farber Cancer Institute, Boston, MA, USA
Rameen Beroukhim & Rameen Beroukhim
Indiana University, Bloomington, IN, USA
S. Cenk Sahinalp & S. Cenk Sahinalp
The University of Chicago, Chicago, IL, USA
Yuan Ji & Yuan Ji
University of Cologne, Cologne, Germany
Martin Peifer, Yupeng Cun, Martin Peifer & Tsun-Po Yang
University of Helsinki, Helsinki, Finland
Ville Mustonen & Ville Mustonen
University of Glasgow, Glasgow, UK
Ke Yuan & Ke Yuan
Oxford NIHR Biomedical Research Centre, Oxford, UK
David C. Wedge & David C. Wedge
Department of Computer Science, Carleton College, Northfield, MN, USA
Layla Oesper
Department of Computer Science, Princeton University, Princeton, NJ, USA
Benjamin J. Raphael
Korea University, Seoul, South Korea
Seung Jun Shin
Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX, USA
David A. Wheeler
Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Helsinki, Finland
- Lauri A. Aaltonen
Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK
Federico Abascal, David J. Adams, Ludmil B. Alexandrov, Sam Behjati, Shriram G. Bhosle, David T. Bowen, Adam P. Butler, Peter J. Campbell, Peter Clapham, Helen Davies, Kevin J. Dawson, Stefan C. Dentro, Serge Serge, Erik Garrison, Mohammed Ghori, Dominik Glodzik, Jonathan Hinton, David R. Jones, Young Seok Ju, Stian Knappskog, Barbara Kremeyer, Henry Lee-Six, Daniel A. Leongamornlert, Yilong Li, Sancha Martin, Iñigo Martincorena, Ultan McDermott, Andrew Menzies, Thomas J. Mitchell, Sandro Morganella, Jyoti Nangalia, Jonathan Nicholson, Serena Nik-Zainal, Sarah O’Meara, Elli Papaemmanuil, Keiran M. Raine, Manasa Ramakrishna, Kamna Ramakrishnan, Nicola D. Roberts, Rebecca Shepherd, Lucy Stebbings, Michael R. Stratton, Maxime Tarabichi, Jon W. Teague, Ignacio Vázquez-García, David C. Wedge, Lucy Yates, Jorge Zamora & Xueqing Zou
Memorial Sloan Kettering Cancer Center, New York, NY, USA
Adam Abeshouse, Hikmat Al-Ahmadie, Gunes Gundem, Zachary Heins, Jason Huse, Douglas A. Levine, Eric Minwei Liu & Angelica Ochoa
Genome Science Division, Research Center for Advanced Science and Technology, University of Tokyo, Tokyo, Japan
Hiroyuki Aburatani, Genta Nagae, Akihiro Suzuki, Kenji Tatsuno & Shogo Yamamoto
Department of Surgery, University of Chicago, Chicago, IL, USA
Nishant Agrawal
Department of Surgery, Division of Hepatobiliary and Pancreatic Surgery, School of Medicine, Keimyung University Dongsan Medical Center, Daegu, South Korea
Keun Soo Ahn & Koo Jeong Kang
Department of Oncology, Gil Medical Center, Gachon University, Incheon, South Korea
Sung-Min Ahn
Hiroshima University, Hiroshima, Japan
Hiroshi Aikata, Koji Arihiro, Kazuaki Chayama, Yoshiiku Kawakami & Hideki Ohdan
Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Rehan Akbani, Shaolong Cao, Yiwen Chen, Zechen Chong, Yu Fan, Jun Li, Han Liang, Wenyi Wang, Yumeng Wang & Yuan Yuan
University of Texas MD Anderson Cancer Center, Houston, TX, USA
Kadir C. Akdemir & Ken Chen
King Faisal Specialist Hospital and Research Centre, Al Maather, Riyadh, Saudi Arabia
Sultan T. Al-Sedairy
Bioinformatics Unit, Spanish National Cancer Research Centre (CNIO), Madrid, Spain
Fatima Al-Shahrour & Elena Piñeiro-Yáñez
Bioinformatics Core Facility, University Medical Center Hamburg, Hamburg, Germany
Malik Alawi
Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany
Malik Alawi & Adam Grundhoff
Ontario Tumour Bank, Ontario Institute for Cancer Research, Toronto, ON, Canada
Monique Albert & John Bartlett
Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Kenneth Aldape, Russell R. Broaddus, Bogdan Czerniak, Adel El-Naggar, Savitri Krishnamurthy, Alexander J. Lazar & Xiaoping Su
Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA
Kenneth Aldape
Department of Cellular and Molecular Medicine and Department of Bioengineering, University of California San Diego, La Jolla, CA, USA
Ludmil B. Alexandrov & Erik N. Bergstrom
UC San Diego Moores Cancer Center, San Diego, CA, USA
Ludmil B. Alexandrov, Erik N. Bergstrom & Olivier Harismendy
Canada’s Michael Smith Genome Sciences Centre, BC Cancer, Vancouver, BC, Canada
Adrian Ally, Miruna Balasundaram, Reanne Bowlby, Denise Brooks, Rebecca Carlsen, Eric Chuah, Noreen Dhalla, Robert A. Holt, Steven J. M. Jones, Katayoon Kasaian, Darlene Lee, Haiyan Irene Li, Yussanne Ma, Marco A. Marra, Michael Mayo, Richard A. Moore, Andrew J. Mungall, Karen Mungall, A. Gordon Robertson, Sara Sadeghi, Jacqueline E. Schein, Payal Sipahimalani, Angela Tam, Nina Thiessen & Tina Wong
Sir Peter MacCallum Department of Oncology, Peter MacCallum Cancer Centre, University of Melbourne, Melbourne, VIC, Australia
Kathryn Alsop, David D. L. Bowtell, Elizabeth L. Christie, Dariush Etemadmoghadam, Sian Fereday, Dale W. Garsed, Linda Mileshkin, Chris Mitchell, Mark Shackleton, Heather Thorne & Nadia Traficante
Centre for Research in Molecular Medicine and Chronic Diseases (CiMUS), Universidade de Santiago de Compostela, Santiago de Compostela, Spain
Eva G. Alvarez, Alicia L. Bruzos, Bernardo Rodriguez-Martin, Javier Temes, Jose M. C. Tubio & Jorge Zamora
Department of Zoology, Genetics and Physical Anthropology, (CiMUS), Universidade de Santiago de Compostela, Santiago de Compostela, Spain
The Biomedical Research Centre (CINBIO), Universidade de Vigo, Vigo, Spain
Eva G. Alvarez, Alicia L. Bruzos, Bernardo Rodriguez-Martin, Marta Tojo, Jose M. C. Tubio & Jorge Zamora
Royal National Orthopaedic Hospital - Bolsover, London, UK
Fernanda Amary
Department of Genomic Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Samirkumar B. Amin, P. Andrew Futreal & Alexander J. Lazar
Quantitative and Computational Biosciences Graduate Program, Baylor College of Medicine, Houston, TX, USA
Samirkumar B. Amin, Han Liang & Yumeng Wang
The Jackson Laboratory for Genomic Medicine, Farmington, CT, USA
Samirkumar B. Amin, Joshy George & Lucas Lochovsky
Genome Informatics Program, Ontario Institute for Cancer Research, Toronto, ON, Canada
Brice Aminou, Niall J. Byrne, Aurélien Chateigner, Nodirjon Fayzullaev, Vincent Ferretti, George L. Mihaiescu, Hardeep K. Nahal-Bose, Brian D. O’Connor, B. F. Francis Ouellette, Marc D. Perry, Kevin Thai, Qian Xiang, Christina K. Yung & Junjun Zhang
Institute of Human Genetics, Christian-Albrechts-University, Kiel, Germany
Ole Ammerpohl, Andrea Haake, Cristina López, Julia Richter & Rabea Wagener
Institute of Human Genetics, Ulm University and Ulm University Medical Center, Ulm, Germany
Ole Ammerpohl, Sietse Aukema, Cristina López, Reiner Siebert & Rabea Wagener
Queensland Centre for Medical Genomics, Institute for Molecular Bioscience, University of Queensland, St. Lucia, Brisbane, QLD, Australia
Matthew J. Anderson, Timothy J. C. Bruxner, Angelika N. Christ, J. Lynn Fink, Ivon Harliwong, Karin S. Kassahn, David K. Miller, Alan J. Robertson & Darrin F. Taylor
Salford Royal NHS Foundation Trust, Salford, UK
Yeng Ang, Hsiao-Wei Chen, Ritika Kundra & Francisco Sanchez-Vega
Department of Surgery, Pancreas Institute, University and Hospital Trust of Verona, Verona, Italy
Davide Antonello, Claudio Bassi, Narong Khuntikeo, Luca Landoni, Giuseppe Malleo, Giovanni Marchegiani, Neil D. Merrett, Marco Miotto, Salvatore Paiella, Antonio Pea, Paolo Pederzoli, Roberto Salvia, Jaswinder S. Samra, Elisabetta Sereni & Samuel Singer
Molecular and Medical Genetics, OHSU Knight Cancer Institute, Oregon Health and Science University, Portland, OR, USA
Pavana Anur, Myron Peto & Paul T. Spellman
Department of Molecular Oncology, BC Cancer Research Centre, Vancouver, BC, Canada
Samuel Aparicio
The McDonnell Genome Institute at Washington University, St. Louis, MO, USA
Elizabeth L. Appelbaum, Matthew H. Bailey, Matthew G. Cordes, Li Ding, Catrina C. Fronick, Lucinda A. Fulton, Robert S. Fulton, Kuan-lin Huang, Reyka Jayasinghe, Elaine R. Mardis, R. Jay Mashl, Michael D. McLellan, Christopher A. Miller, Heather K. Schmidt, Jiayin Wang, Michael C. Wendl, Richard K. Wilson & Tina Wong
University College London, London, UK
Elizabeth L. Appelbaum, Jonathan D. Kay, Helena Kilpinen, Laurence B. Lovat, Hayley J. Luxton & Hayley C. Whitaker
Division of Cancer Genomics, National Cancer Center Research Institute, National Cancer Center, Tokyo, Japan
Yasuhito Arai, Natsuko Hama, Fumie Hosoda, Hiromi Nakamura, Tatsuhiro Shibata, Yasushi Totoki & Shinichi Yachida
DLR Project Management Agency, Bonn, Germany
Tokyo Women’s Medical University, Tokyo, Japan
Shun-ichi Ariizumi & Masakazu Yamamoto
Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Joshua Armenia, Hsiao-Wei Chen, Jianjiong Gao, Ritika Kundra, Francisco Sanchez-Vega, Nikolaus Schultz & Hongxin Zhang
Los Alamos National Laboratory, Los Alamos, NM, USA
Laurent Arnould
Department of Pathology, University Health Network, Toronto General Hospital, Toronto, ON, Canada
Sylvia Asa, Michael H. A. Roehrl & Theodorus Van der Kwast
Nottingham University Hospitals NHS Trust, Nottingham, UK
Sylvia Asa, Simon L. Parsons & Ming Tsao
Epigenomics and Cancer Risk Factors, German Cancer Research Center (DKFZ), Heidelberg, Germany
Yassen Assenov
Computational Biology Program, Ontario Institute for Cancer Research, Toronto, ON, Canada
Gurnit Atwal, Philip Awadalla, Jonathan Barenboim, Vinayak Bhandari, Ivan Borozan, Paul C. Boutros, Lewis Jonathan Dursi, Shadrielle M. G. Espiritu, Natalie S. Fox, Michael Fraser, Syed Haider, Vincent Huang, Keren Isaev, Wei Jiao, Christopher M. Lalansingh, Emilie Lalonde, Fabien C. Lamaze, Constance H. Li, Julie Livingstone, Christine P’ng, Marta Paczkowska, Stephenie D. Prokopec, Jüri Reimand, Veronica Y. Sabelnykova, Adriana Salcedo, Yu-Jia Shiah, Solomon I. Shorser, Shimin Shuai, Jared T. Simpson, Lincoln D. Stein, Ren X. Sun, Lina Wadi, Gavin W. Wilson, Adam J. Wright, Takafumi N. Yamaguchi, Fouad Yousif & Denis Yuen
Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada
Gurnit Atwal, Philip Awadalla, Gary D. Bader, Shimin Shuai & Lincoln D. Stein
Vector Institute, Toronto, ON, Canada
Gurnit Atwal, Quaid D. Morris, Yulia Rubanova & Jeffrey A. Wintersinger
Hematopathology Section, Institute of Pathology, Christian-Albrechts-University, Kiel, Germany
Sietse Aukema, Wolfram Klapper, Julia Richter & Monika Szczepanowski
Department of Pathology and Laboratory Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
J. Todd Auman & Charles M. Perou
Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital, The Norwegian Radium Hospital, Oslo, Norway
Miriam R. R. Aure, Anne-Lise Børresen-Dale & Anita Langerød
Pathology, Hospital Clinic, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), University of Barcelona, Barcelona, Spain
Marta Aymerich
Department of Veterinary Medicine, Transmissible Cancer Group, University of Cambridge, Cambridge, UK
Adrian Baez-Ortega
Alvin J. Siteman Cancer Center, Washington University School of Medicine, St. Louis, MO, USA
Matthew H. Bailey, Li Ding, Robert S. Fulton, Ramaswamy Govindan & Michael D. McLellan
Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Glasgow, UK
Peter J. Bailey, Andrew V. Biankin, David K. Chang, Susanna L. Cooke, Fraser R. Duthie, Janet S. Graham, Nigel B. Jamieson, Elizabeth A. Musgrove & Derek W. Wright
Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Saianand Balu, Tom Bodenheimer, D. Neil Hayes, Austin J. Hepperla, Katherine A. Hoadley, Alan P. Hoyle, Stuart R. Jefferys, Shaowu Meng, Lisle E. Mose, Grant Sanders, Yan Shi, Janae V. Simons & Matthew G. Soloway
Pratiti Bandopadhayay, Rameen Beroukhim, Angela N. Brooks, Susan Bullman, John Busanovich, Andrew D. Cherniack, Juok Cho, Carrie Cibulskis, Kristian Cibulskis, David Craft, Timothy Defreitas, Andrew J. Dunford, Scott Frazer, Stacey B. Gabriel, Nils Gehlenborg, Gad Getz, Manaswi Gupta, Gavin Ha, Nicholas J. Haradhvala, David I. Heiman, Julian M. Hess, Manolis Kellis, Jaegil Kim, Kiran Kumar, Kirsten Kübler, Eric Lander, Michael S. Lawrence, Ignaty Leshchiner, Pei Lin, Ziao Lin, Dimitri Livitz, Yosef E. Maruvka, Samuel R. Meier, Matthew Meyerson, Michael S. Noble, Chandra Sekhar Pedamallu, Paz Polak, Esther Rheinbay, Daniel Rosebrock, Mara Rosenberg, Gordon Saksena, Richard Sallari, Steven E. Schumacher, Ayellet V. Segre, Ofer Shapira, Juliann Shih, Nasa Sinnott-Armstrong, Oliver Spiro, Chip Stewart, Amaro Taylor-Weiner, Grace Tiao, Douglas Voet, Jeremiah A. Wala, Cheng-Zhong Zhang & Hailei Zhang
Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Boston, MA, USA
Pratiti Bandopadhayay
Department of Pediatrics, Harvard Medical School, Boston, MA, USA
Leeds Institute of Medical Research @ St. James’s, University of Leeds, St. James’s University Hospital, Leeds, UK
Rosamonde E. Banks & Naveen Vasudev
Department of Pathology and Diagnostics, University and Hospital Trust of Verona, Verona, Italy
Stefano Barbi, Vincenzo Corbo & Michele Simbolo
Department of Surgery, Princess Alexandra Hospital, Brisbane, QLD, Australia
Andrew P. Barbour
Surgical Oncology Group, Diamantina Institute, University of Queensland, Brisbane, QLD, Australia
Department of Population and Quantitative Health Sciences, Case Western Reserve University School of Medicine, Cleveland, OH, USA
Jill Barnholtz-Sloan
Research Health Analytics and Informatics, University Hospitals Cleveland Medical Center, Cleveland, OH, USA
Gloucester Royal Hospital, Gloucester, UK
Elisabet Barrera, Wojciech Bazant, Ewan Birney, Rich Boyce, Alvis Brazma, Andy Cafferkey, Claudia Calabrese, Paul Flicek, Nuno A. Fonseca, Anja Füllgrabe, Moritz Gerstung, Santiago Gonzalez, Liliana Greger, Maria Keays, Jan O. Korbel, Alfonso Muñoz, Steven J. Newhouse, David Ocana, Irene Papatheodorou, Robert Petryszak, Roland F. Schwarz, Charles Short, Oliver Stegle & Lara Urban
Diagnostic Development, Ontario Institute for Cancer Research, Toronto, ON, Canada
John Bartlett & Ilinca Lungu
Barcelona Supercomputing Center (BSC), Barcelona, Spain
Javier Bartolome, Mattia Bosio, Ana Dueso-Barroso, J. Lynn Fink, Josep L. L. Gelpi, Ana Milovanovic, Montserrat Puiggròs, Javier Bartolomé Rodriguez, Romina Royo, David Torrents, Alfonso Valencia, Miguel Vazquez, David Vicente & Izar Villasante
Arnie Charbonneau Cancer Institute, University of Calgary, Calgary, AB, Canada
Oliver F. Bathe
Departments of Surgery and Oncology, University of Calgary, Calgary, AB, Canada
Department of Pathology, Oslo University Hospital, The Norwegian Radium Hospital, Oslo, Norway
Daniel Baumhoer & Bodil Bjerkehagen
PanCuRx Translational Research Initiative, Ontario Institute for Cancer Research, Toronto, ON, Canada
Prashant Bavi, Michelle Chan-Seng-Yue, Sean Cleary, Robert E. Denroche, Steven Gallinger, Robert C. Grant, Gun Ho Jang, Sangeetha Kalimuthu, Ilinca Lungu, John D. McPherson, Faiyaz Notta, Michael H. A. Roehrl, Gavin W. Wilson & Julie M. Wilson
Department of Oncology, Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University School of Medicine, Baltimore, MD, USA
Stephen B. Baylin, Nilanjan Chatterjee, Leslie Cope, Ludmila Danilova & Ralph H. Hruban
University Hospital Southampton NHS Foundation Trust, Southampton, UK
Stephen B. Baylin & Tim Dudderidge
Royal Stoke University Hospital, Stoke-on-Trent, UK
Duncan Beardsmore & Christopher Umbricht
Genome Sequence Informatics, Ontario Institute for Cancer Research, Toronto, ON, Canada
Timothy A. Beck, Bob Gibson, Lawrence E. Heisler, Xuemei Luo & Morgan L. Taschuk
Human Longevity Inc, San Diego, CA, USA
Timothy A. Beck
Olivia Newton-John Cancer Research Institute, La Trobe University, Heidelberg, VIC, Australia
Andreas Behren & Jonathan Cebon
Computer Network Information Center, Chinese Academy of Sciences, Beijing, China
Beifang Niu
Genome Canada, Ottawa, ON, Canada
CNAG-CRG, Centre for Genomic Regulation (CRG), Barcelona Institute of Science and Technology (BIST), Barcelona, Spain
Sergi Beltran, Ivo G. Gut, Marta Gut, Simon C. Heath, Tomas Marques-Bonet, Arcadi Navarro, Miranda D. Stobbe, Jean-Rémi Trotta & Justin P. Whalley
Universitat Pompeu Fabra (UPF), Barcelona, Spain
Sergi Beltran, Mattia Bosio, German M. Demidov, Oliver Drechsel, Ivo G. Gut, Marta Gut, Simon C. Heath, Francesc Muyas, Stephan Ossowski, Aparna Prasad, Raquel Rabionet, Miranda D. Stobbe & Hana Susak
Buck Institute for Research on Aging, Novato, CA, USA
Christopher Benz & Christina Yau
Duke University Medical Center, Durham, NC, USA
Andrew Berchuck
Department of Human Genetics, Hannover Medical School, Hannover, Germany
Anke K. Bergmann
Center for Bioinformatics and Functional Genomics, Cedars-Sinai Medical Center, Los Angeles, CA, USA
Benjamin P. Berman & Huy Q. Dinh
Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA
Benjamin P. Berman
The Hebrew University Faculty of Medicine, Jerusalem, Israel
Barts Cancer Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, UK
Daniel M. Berney & Yong-Jie Lu
Department of Computer Science, Bioinformatics Group, University of Leipzig, Leipzig, Germany
Stephan H. Bernhart, Hans Binder, Steve Hoffmann & Peter F. Stadler
Interdisciplinary Center for Bioinformatics, University of Leipzig, Leipzig, Germany
Stephan H. Bernhart, Hans Binder, Steve Hoffmann, Helene Kretzmer & Peter F. Stadler
Transcriptome Bioinformatics, LIFE Research Center for Civilization Diseases, University of Leipzig, Leipzig, Germany
Stephan H. Bernhart, Steve Hoffmann, Helene Kretzmer & Peter F. Stadler
Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA
Rameen Beroukhim, Angela N. Brooks, Susan Bullman, Andrew D. Cherniack, Levi Garraway, Matthew Meyerson, Chandra Sekhar Pedamallu, Steven E. Schumacher, Juliann Shih & Jeremiah A. Wala
Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA
Rameen Beroukhim, Aquila Fatima, Andrea L. Richardson, Steven E. Schumacher, Ofer Shapira, Andrew Tutt & Jeremiah A. Wala
Rameen Beroukhim, Gad Getz, Kirsten Kübler, Matthew Meyerson, Chandra Sekhar Pedamallu, Paz Polak, Esther Rheinbay & Jeremiah A. Wala
USC Norris Comprehensive Cancer Center, University of Southern California, Los Angeles, CA, USA
Mario Berrios, Moiz S. Bootwalla, Andrea Holbrook, Phillip H. Lai, Dennis T. Maglinte, David J. Van Den Berg & Daniel J. Weisenberger
Department of Diagnostics and Public Health, University and Hospital Trust of Verona, Verona, Italy
Samantha Bersani, Ivana Cataldo, Claudio Luchini & Maria Scardoni
Department of Mathematics, Aarhus University, Aarhus, Denmark
Johanna Bertl & Asger Hobolth
Department of Molecular Medicine (MOMA), Aarhus University Hospital, Aarhus N, Denmark
Johanna Bertl, Henrik Hornshøj, Malene Juul, Randi Istrup Juul, Tobias Madsen, Morten Muhlig Nielsen & Jakob Skou Pedersen
Instituto Carlos Slim de la Salud, Mexico City, Mexico
Miguel Betancourt
Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada
Vinayak Bhandari, Paul C. Boutros, Robert G. Bristow, Keren Isaev, Constance H. Li, Jüri Reimand, Michael H. A. Roehrl & Bradly G. Wouters
Cancer Division, Garvan Institute of Medical Research, Kinghorn Cancer Centre, University of New South Wales (UNSW Sydney), Sydney, NSW, Australia
Andrew V. Biankin, David K. Chang, Lorraine A. Chantrill, Angela Chou, Anthony J. Gill, Amber L. Johns, James G. Kench, David K. Miller, Adnan M. Nagrial, Marina Pajic, Mark Pinese, Ilse Rooman, Christopher J. Scarlett, Christopher W. Toon & Jianmin Wu
South Western Sydney Clinical School, Faculty of Medicine, University of New South Wales (UNSW Sydney), Liverpool, NSW, Australia
Andrew V. Biankin
West of Scotland Pancreatic Unit, Glasgow Royal Infirmary, Glasgow, UK
Andrew V. Biankin & Nigel B. Jamieson
Center for Digital Health, Berlin Institute of Health and Charitè - Universitätsmedizin Berlin, Berlin, Germany
Matthias Bieg
Heidelberg Center for Personalized Oncology (DKFZ-HIPO), German Cancer Research Center (DKFZ), Heidelberg, Germany
Matthias Bieg, Ivo Buchhalter, Barbara Hutter & Nagarajan Paramasivam
The Preston Robert Tisch Brain Tumor Center, Duke University Medical Center, Durham, NC, USA
Darell Bigner
Massachusetts General Hospital, Boston, MA, USA
Michael Birrer, Vikram Deshpande, William C. Faquin, Nicholas J. Haradhvala, Kirsten Kübler, Michael S. Lawrence, David N. Louis, Yosef E. Maruvka, G. Petur Nielsen, Esther Rheinbay, Mara Rosenberg, Dennis C. Sgroi & Chin-Lee Wu
National Institute of Biomedical Genomics, Kalyani, West Bengal, India
Nidhan K. Biswas, Arindam Maitra & Partha P. Majumder
Institute of Clinical Medicine and Institute of Oral Biology, University of Oslo, Oslo, Norway
Bodil Bjerkehagen
University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Lori Boice, Mei Huang, Sonia Puig & Leigh B. Thorne
ARC-Net Centre for Applied Research on Cancer, University and Hospital Trust of Verona, Verona, Italy
Giada Bonizzato, Cinzia Cantù, Ivana Cataldo, Vincenzo Corbo, Sonia Grimaldi, Rita T. Lawlor, Andrea Mafficini, Borislav C. Rusev, Aldo Scarpa, Katarzyna O. Sikora, Nicola Sperandio, Alain Viari & Caterina Vicentini
The Institute of Cancer Research, London, UK
Johann S. De Bono, Niedzica Camacho, Colin S. Cooper, Sandra E. Edwards, Rosalind A. Eeles, Zsofia Kote-Jarai, Daniel A. Leongamornlert, Lucy Matthews & Sue Merson
Centre for Computational Biology, Duke-NUS Medical School, Singapore, Singapore
Arnoud Boot, Ioana Cutcutache, Mi Ni Huang, John R. McPherson, Steven G. Rozen & Yang Wu
Programme in Cancer and Stem Cell Biology, Duke-NUS Medical School, Singapore, Singapore
Arnoud Boot, Ioana Cutcutache, Mi Ni Huang, John R. McPherson, Steven G. Rozen, Patrick Tan, Bin Tean Teh & Yang Wu
Division of Oncology and Pathology, Department of Clinical Sciences Lund, Lund University, Lund, Sweden
Ake Borg, Markus Ringnér & Johan Staaf
Department of Pediatric Oncology, Hematology and Clinical Immunology, Heinrich-Heine-University, Düsseldorf, Germany
Arndt Borkhardt & Jessica I. Hoell
Laboratory for Medical Science Mathematics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
Keith A. Boroevich, Todd A. Johnson, Michael S. Lawrence & Tatsuhiko Tsunoda
RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
Keith A. Boroevich, Akihiro Fujimoto, Masashi Fujita, Mayuko Furuta, Kazuhiro Maejima, Hidewaki Nakagawa, Kaoru Nakano & Aya Sasaki-Oku
Department of Internal Medicine/Hematology, Friedrich-Ebert-Hospital, Neumünster, Germany
Christoph Borst & Siegfried Haas
Departments of Dermatology and Pathology, Yale University, New Haven, CT, USA
Marcus Bosenberg
Centre for Genomic Regulation (CRG), The Barcelona Institute of Science and Technology, Barcelona, Spain
Mattia Bosio, German M. Demidov, Oliver Drechsel, Georgia Escaramis, Xavier Estivill, Aliaksei Z. Holik, Francesc Muyas, Stephan Ossowski, Raquel Rabionet & Hana Susak
Radcliffe Department of Medicine, University of Oxford, Oxford, UK
Jacqueline Boultwood
Canadian Center for Computational Genomics, McGill University, Montreal, QC, Canada
Guillaume Bourque
Department of Human Genetics, McGill University, Montreal, QC, Canada
Guillaume Bourque, Mark Lathrop & Yasser Riazalhosseini
Department of Human Genetics, University of California Los Angeles, Los Angeles, CA, USA
Paul C. Boutros
Department of Pharmacology, University of Toronto, Toronto, ON, Canada
Faculty of Medicine and Health Technology, Tampere University and Tays Cancer Center, Tampere University Hospital, Tampere, Finland
G. Steven Bova & Tapio Visakorpi
Haematology, Leeds Teaching Hospitals NHS Trust, Leeds, UK
David T. Bowen
Translational Research and Innovation, Centre Léon Bérard, Lyon, France
Sandrine Boyault
Fox Chase Cancer Center, Philadelphia, PA, USA
Jeffrey Boyd & Elaine R. Mardis
International Agency for Research on Cancer, World Health Organization, Lyon, France
Paul Brennan & Ghislaine Scelo
Earlham Institute, Norwich, UK
Daniel S. Brewer & Colin S. Cooper
Norwich Medical School, University of East Anglia, Norwich, UK
Department of Molecular Biology, Faculty of Science, Radboud Institute for Molecular Life Sciences, Radboud University, Nijmegen, HB, The Netherlands
Arie B. Brinkman
CRUK Manchester Institute and Centre, Manchester, UK
Robert G. Bristow
Department of Radiation Oncology, University of Toronto, Toronto, ON, Canada
Division of Cancer Sciences, Manchester Cancer Research Centre, University of Manchester, Manchester, UK
Radiation Medicine Program, Princess Margaret Cancer Centre, Toronto, ON, Canada
Robert G. Bristow & Fei-Fei Fei Liu
Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA
Jane E. Brock & Sabina Signoretti
Department of Surgery, Division of Thoracic Surgery, The Johns Hopkins University School of Medicine, Baltimore, MD, USA
Malcolm Brock
Division of Molecular Pathology, The Netherlands Cancer Institute, Oncode Institute, Amsterdam, CX, The Netherlands
Annegien Broeks & Jos Jonkers
Department of Biomolecular Engineering, University of California Santa Cruz, Santa Cruz, CA, USA
Angela N. Brooks, David Haan, Maximillian G. Marin, Thomas J. Matthew, Yulia Newton, Cameron M. Soulette & Joshua M. Stuart
UC Santa Cruz Genomics Institute, University of California Santa Cruz, Santa Cruz, CA, USA
Angela N. Brooks, Brian Craft, Mary J. Goldman, David Haussler, Joshua M. Stuart & Jingchun Zhu
Division of Applied Bioinformatics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Benedikt Brors, Lars Feuerbach, Chen Hong, Charles David Imbusch & Lina Sieverling
German Cancer Consortium (DKTK), German Cancer Research Center (DKFZ), Heidelberg, Germany
Benedikt Brors, Barbara Hutter, Peter Lichter, Dirk Schadendorf & Holger Sültmann
National Center for Tumor Diseases (NCT) Heidelberg, Heidelberg, Germany
Benedikt Brors, Barbara Hutter, Holger Sültmann & Thorsten Zenz
Center for Biological Sequence Analysis, Department of Bio and Health Informatics, Technical University of Denmark, Lyngby, Denmark
Søren Brunak
Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, Copenhagen, Denmark
Institute for Molecular Bioscience, University of Queensland, St. Lucia, Brisbane, QLD, Australia
Timothy J. C. Bruxner, Oliver Holmes, Stephen H. Kazakoff, Conrad R. Leonard, Felicity Newell, Katia Nones, Ann-Marie Patch, John V. Pearson, Michael C. Quinn, Nick M. Waddell, Nicola Waddell, Scott Wood & Qinying Xu
Biomedical Engineering, Oregon Health and Science University, Portland, OR, USA
Alex Buchanan & Kyle Ellrott
Division of Theoretical Bioinformatics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Ivo Buchhalter, Calvin Wing Yiu Chan, Roland Eils, Michael C. Heinold, Carl Herrmann, Natalie Jäger, Rolf Kabbe, Jules N. A. Kerssemakers, Kortine Kleinheinz, Nagarajan Paramasivam, Manuel Prinz, Matthias Schlesner & Johannes Werner
Institute of Pharmacy and Molecular Biotechnology and BioQuant, Heidelberg University, Heidelberg, Germany
Ivo Buchhalter, Roland Eils, Michael C. Heinold, Carl Herrmann, Daniel Hübschmann, Kortine Kleinheinz & Umut H. Toprak
Federal Ministry of Education and Research, Berlin, Germany
Christiane Buchholz
Melanoma Institute Australia, University of Sydney, Sydney, NSW, Australia
Hazel Burke, Ricardo De Paoli-Iseppi, Nicholas K. Hayward, Peter Hersey, Valerie Jakrot, Hojabr Kakavand, Georgina V. Long, Graham J. Mann, Robyn P. M. Saw, Richard A. Scolyer, Ping Shang, Andrew J. Spillane, Jonathan R. Stretch, John F. F. Thompson & James S. Wilmott
Pediatric Hematology and Oncology, University Hospital Muenster, Muenster, Germany
Birgit Burkhardt
Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD, USA
Kathleen H. Burns & Christopher Umbricht
McKusick-Nathans Institute of Genetic Medicine, Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University School of Medicine, Baltimore, MD, USA
Kathleen H. Burns
Foundation Medicine, Inc, Cambridge, MA, USA
John Busanovich
Department of Biomedical Data Science, Stanford University School of Medicine, Stanford, CA, USA
Carlos D. Bustamante & Francisco M. De La Vega
Department of Genetics, Stanford University School of Medicine, Stanford, CA, USA
Carlos D. Bustamante, Francisco M. De La Vega, Suyash S. Shringarpure, Nasa Sinnott-Armstrong & Mark H. Wright
Bakar Computational Health Sciences Institute and Department of Pediatrics, University of California, San Francisco, CA, USA
Atul J. Butte & Jieming Chen
Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway
Anne-Lise Børresen-Dale
National Cancer Institute, National Institutes of Health, Bethesda, MD, USA
Samantha J. Caesar-Johnson, John A. Demchok, Ina Felau, Roy Tarnuzzer, Zhining Wang, Liming Yang, Jean C. Zenklusen & Jiashan Zhang
Royal Marsden NHS Foundation Trust, London and Sutton, UK
Declan Cahill, Nening M. Dennis, Tim Dudderidge, Rosalind A. Eeles, Cyril Fisher, Steven Hazell, Vincent Khoo, Pardeep Kumar, Naomi Livni, Erik Mayer, David Nicol, Christopher Ogden, Edward W. Rowe, Sarah Thomas, Alan Thompson & Nicholas van As
Genome Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany
Claudia Calabrese, Serap Erkek, Moritz Gerstung, Santiago Gonzalez, Nina Habermann, Wolfgang Huber, Lara Jerman, Jan O. Korbel, Esa Pitkänen, Benjamin Raeder, Tobias Rausch, Vasilisa A. Rudneva, Oliver Stegle, Stephanie Sungalee, Lara Urban, Sebastian M. Waszak, Joachim Weischenfeldt & Sergei Yakneen
Department of Oncology, University of Cambridge, Cambridge, UK
Carlos Caldas & Suet-Feung Chin
Li Ka Shing Centre, Cancer Research UK Cambridge Institute, University of Cambridge, Cambridge, UK
Carlos Caldas, Suet-Feung Chin, Ruben M. Drews, Paul A. Edwards, Matthew Eldridge, Steve Hawkins, Andy G. Lynch, Geoff Macintyre, Florian Markowetz, Charlie E. Massie, David E. Neal, Simon Tavaré & Ke Yuan
Institut Gustave Roussy, Villejuif, France
Fabien Calvo
Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK
Peter J. Campbell, Vincent J. Gnanapragasam, William Howat, Thomas J. Mitchell, David E. Neal, Nimish C. Shah & Anne Y. Warren
Department of Haematology, University of Cambridge, Cambridge, UK
Peter J. Campbell
Anatomia Patológica, Hospital Clinic, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), University of Barcelona, Barcelona, Spain
Elias Campo
Spanish Ministry of Science and Innovation, Madrid, Spain
University of Michigan Comprehensive Cancer Center, Ann Arbor, MI, USA
Thomas E. Carey
Department for BioMedical Research, University of Bern, Bern, Switzerland
Joana Carlevaro-Fita
Department of Medical Oncology, Inselspital, University Hospital and University of Bern, Bern, Switzerland
Joana Carlevaro-Fita, Rory Johnson & Andrés Lanzós
Graduate School for Cellular and Biomedical Sciences, University of Bern, Bern, Switzerland
Joana Carlevaro-Fita & Andrés Lanzós
University of Pavia, Pavia, Italy
Mario Cazzola & Luca Malcovati
University of Alabama at Birmingham, Birmingham, AL, USA
Robert Cerfolio
UHN Program in BioSpecimen Sciences, Toronto General Hospital, Toronto, ON, Canada
Dianne E. Chadwick, Sheng-Ben Liang, Michael H. A. Roehrl & Sagedeh Shahabi
Department of Urology, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Dimple Chakravarty
Centre for Law and Genetics, University of Tasmania, Sandy Bay Campus, Hobart, TAS, Australia
Don Chalmers
Faculty of Biosciences, Heidelberg University, Heidelberg, Germany
Calvin Wing Yiu Chan, Chen Hong & Lina Sieverling
Department of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Ottawa, ON, Canada
Division of Anatomic Pathology, Mayo Clinic, Rochester, MN, USA
Vishal S. Chandan
Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA
Stephen J. Chanock, Xing Hua, Lisa Mirabello, Lei Song & Bin Zhu
Illawarra Shoalhaven Local Health District L3 Illawarra Cancer Care Centre, Wollongong Hospital, Wollongong, NSW, Australia
Lorraine A. Chantrill
BioForA, French National Institute for Agriculture, Food, and Environment (INRAE), ONF, Orléans, France
Aurélien Chateigner
Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, USA
Nilanjan Chatterjee
University of California San Diego, San Diego, CA, USA
Zhaohong Chen, Michelle T. Dow, Claudiu Farcas, S. M. Ashiqul Islam, Antonios Koures, Lucila Ohno-Machado, Christos Sotiriou & Ashley Williams
Division of Experimental Pathology, Mayo Clinic, Rochester, MN, USA
Jeremy Chien
Centre for Cancer Research, The Westmead Institute for Medical Research, University of Sydney, Sydney, NSW, Australia
Yoke-Eng Chiew, Angela Chou, Jillian A. Hung, Catherine J. Kennedy, Graham J. Mann, Gulietta M. Pupo, Sarah-Jane Schramm, Varsha Tembe & Anna deFazio
Department of Gynaecological Oncology, Westmead Hospital, Sydney, NSW, Australia
Yoke-Eng Chiew, Jillian A. Hung, Catherine J. Kennedy & Anna deFazio
PDXen Biosystems Inc, Seoul, South Korea
Sunghoon Cho
Korea Advanced Institute of Science and Technology, Daejeon, South Korea
Jung Kyoon Choi, Young Seok Ju & Christopher J. Yoon
Electronics and Telecommunications Research Institute, Daejeon, South Korea
Wan Choi, Seung-Hyup Jeon, Hyunghwan Kim & Youngchoon Woo
Institut National du Cancer (INCA), Boulogne-Billancourt, France
Christine Chomienne & Iris Pauporté
Department of Genetics, Informatics Institute, University of Alabama at Birmingham, Birmingham, AL, USA
Zechen Chong
Division of Medical Oncology, National Cancer Centre, Singapore, Singapore
Su Pin Choo
Medical Oncology, University and Hospital Trust of Verona, Verona, Italy
Sara Cingarlini & Michele Milella
Department of Pediatrics, University Hospital Schleswig-Holstein, Kiel, Germany
Alexander Claviez
Hepatobiliary/Pancreatic Surgical Oncology Program, University Health Network, Toronto, ON, Canada
Sean Cleary, Ashton A. Connor & Steven Gallinger
School of Biological Sciences, University of Auckland, Auckland, New Zealand
Nicole Cloonan
Department of Surgery, University of Melbourne, Parkville, VIC, Australia
Marek Cmero
The Murdoch Children’s Research Institute, Royal Children’s Hospital, Parkville, VIC, Australia
Walter and Eliza Hall Institute, Parkville, VIC, Australia
Vancouver Prostate Centre, Vancouver, Canada
Colin C. Collins, Nilgun Donmez, Faraz Hach, Salem Malikic, S. Cenk Sahinalp, Iman Sarrafi & Raunak Shrestha
Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, ON, Canada
Ashton A. Connor, Steven Gallinger, Robert C. Grant, Treasa A. McPherson & Iris Selander
University of East Anglia, Norwich, UK
Colin S. Cooper
Norfolk and Norwich University Hospital NHS Trust, Norwich, UK
Matthew G. Cordes, Catrina C. Fronick & Tom Roques
Victorian Institute of Forensic Medicine, Southbank, VIC, Australia
Stephen M. Cordner
Department of Biomedical Informatics, Harvard Medical School, Boston, MA, USA
Isidro Cortés-Ciriano, Jake June-Koo Lee & Peter J. Park
Department of Chemistry, Centre for Molecular Science Informatics, University of Cambridge, Cambridge, UK
Isidro Cortés-Ciriano
Ludwig Center at Harvard Medical School, Boston, MA, USA
Kyle Covington, HarshaVardhan Doddapaneni, Richard A. Gibbs, Jianhong Hu, Joy C. Jayaseelan, Viktoriya Korchina, Lora Lewis, Donna M. Muzny, Linghua Wang, David A. Wheeler & Liu Xi
Peter MacCallum Cancer Centre, University of Melbourne, Melbourne, VIC, Australia
Prue A. Cowin, Anne Hamilton, Gisela Mir Arnau & Ravikiran Vedururu
Physics Division, Optimization and Systems Biology Lab, Massachusetts General Hospital, Boston, MA, USA
David Craft
Department of Medicine, Baylor College of Medicine, Houston, TX, USA
Chad J. Creighton
Yupeng Cun, Martin Peifer & Tsun-Po Yang
International Genomics Consortium, Phoenix, AZ, USA
Erin Curley & Troy Shelton
Genomics Research Program, Ontario Institute for Cancer Research, Toronto, ON, Canada
Karolina Czajka, Jenna Eagles, Thomas J. Hudson, Jeremy Johns, Faridah Mbabaali, John D. McPherson, Jessica K. Miller, Danielle Pasternack, Michelle Sam & Lee E. Timms
Barking Havering and Redbridge University Hospitals NHS Trust, Romford, UK
Bogdan Czerniak, Adel El-Naggar & David Khoo
Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia
Rebecca A. Dagg
Department of Medicine, Section of Endocrinology, University and Hospital Trust of Verona, Verona, Italy
Maria Vittoria Davi
Computational Biology Center, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Natalie R. Davidson, Andre Kahles, Kjong-Van Lehmann, Alessandro Pastore, Gunnar Rätsch, Chris Sander, Yasin Senbabaoglu & Nicholas D. Socci
Department of Biology, ETH Zurich, Zürich, Switzerland
Natalie R. Davidson, Andre Kahles, Kjong-Van Lehmann, Gunnar Rätsch & Stefan G. Stark
Department of Computer Science, ETH Zurich, Zurich, Switzerland
Natalie R. Davidson, Andre Kahles, Kjong-Van Lehmann & Gunnar Rätsch
SIB Swiss Institute of Bioinformatics, Lausanne, Switzerland
Weill Cornell Medical College, New York, NY, USA
Natalie R. Davidson, Bishoy M. Faltas & Gunnar Rätsch
Academic Department of Medical Genetics, University of Cambridge, Addenbrooke’s Hospital, Cambridge, UK
Helen Davies & Serena Nik-Zainal
MRC Cancer Unit, University of Cambridge, Cambridge, UK
Helen Davies, Rebecca C. Fitzgerald, Nicola Grehan, Serena Nik-Zainal & Maria O’Donovan
Departments of Pediatrics and Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Ian J. Davis
Seven Bridges Genomics, Charlestown, MA, USA
Brandi N. Davis-Dusenbery, Sinisa Ivkovic, Milena Kovacevic, Ana Mijalkovic Lazic, Sanja Mijalkovic, Mia Nastic, Petar Radovic & Nebojsa Tijanic
Annai Systems, Inc, Carlsbad, CA, USA
Francisco M. De La Vega, Tal Shmaya & Dai-Ying Wu
Department of Pathology, General Hospital of Treviso, Department of Medicine, University of Padua, Treviso, Italy
Angelo P. Dei Tos
Department of Computational Biology, University of Lausanne, Lausanne, Switzerland
Olivier Delaneau
Department of Genetic Medicine and Development, University of Geneva Medical School, Geneva, CH, Switzerland
Swiss Institute of Bioinformatics, University of Geneva, Geneva, CH, Switzerland
Jonas Demeulemeester, Stefan C. Dentro, Matthew W. Fittall, Kerstin Haase, Clemency Jolly, Maxime Tarabichi & Peter Van Loo
Jonas Demeulemeester & Peter Van Loo
Institute of Medical Genetics and Applied Genomics, University of Tübingen, Tübingen, Germany
German M. Demidov, Francesc Muyas & Stephan Ossowski
Computational and Systems Biology, Genome Institute of Singapore, Singapore, Singapore
Deniz Demircioğlu & Jonathan Göke
School of Computing, National University of Singapore, Singapore, Singapore
Deniz Demircioğlu
Big Data Institute, Li Ka Shing Centre, University of Oxford, Oxford, UK
Stefan C. Dentro & David C. Wedge
Biomedical Data Science Laboratory, Francis Crick Institute, London, UK
Nikita Desai
Bioinformatics Group, Department of Computer Science, University College London, London, UK
The Edward S. Rogers Sr. Department of Electrical and Computer Engineering, University of Toronto, Toronto, ON, Canada
Amit G. Deshwar
Breast Cancer Translational Research Laboratory JC Heuson, Institut Jules Bordet, Brussels, Belgium
Christine Desmedt
Department of Oncology, Laboratory for Translational Breast Cancer Research, KU Leuven, Leuven, Belgium
Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain
Jordi Deu-Pons, Joan Frigola, Abel Gonzalez-Perez, Ferran Muiños, Loris Mularoni, Oriol Pich, Iker Reyes-Salazar, Carlota Rubio-Perez, Radhakrishnan Sabarinathan & David Tamborero
Research Program on Biomedical Informatics, Universitat Pompeu Fabra, Barcelona, Spain
Jordi Deu-Pons, Abel Gonzalez-Perez, Ferran Muiños, Loris Mularoni, Oriol Pich, Carlota Rubio-Perez, Radhakrishnan Sabarinathan & David Tamborero
Division of Medical Oncology, Princess Margaret Cancer Centre, Toronto, ON, Canada
Neesha C. Dhani, David Hedley & Malcolm J. Moore
Department of Physiology and Biophysics, Weill Cornell Medicine, New York, NY, USA
Priyanka Dhingra, Ekta Khurana, Eric Minwei Liu & Alexander Martinez-Fundichely
Institute for Computational Biomedicine, Weill Cornell Medicine, New York, NY, USA
Department of Pathology, UPMC Shadyside, Pittsburgh, PA, USA
Independent Consultant, Wellesley, USA
Anthony DiBiase
Department of Cell and Molecular Biology, Science for Life Laboratory, Uppsala University, Uppsala, Sweden
Klev Diamanti, Jan Komorowski & Husen M. Umer
Department of Medicine and Department of Genetics, Washington University School of Medicine, St. Louis, St. Louis, MO, USA
Li Ding, Robert S. Fulton, Michael D. McLellan, Michael C. Wendl & Venkata D. Yellapantula
Hefei University of Technology, Anhui, China
Shuai Ding & Shanlin Yang
Translational Cancer Research Unit, GZA Hospitals St.-Augustinus, Center for Oncological Research, Faculty of Medicine and Health Sciences, University of Antwerp, Antwerp, Belgium
Luc Dirix, Steven Van Laere, Gert G. Van den Eynden & Peter Vermeulen
Simon Fraser University, Burnaby, BC, Canada
Nilgun Donmez, Ermin Hodzic, Salem Malikic, S. Cenk Sahinalp & Iman Sarrafi
University of Pennsylvania, Philadelphia, PA, USA
Ronny Drapkin
Faculty of Science and Technology, University of Vic—Central University of Catalonia (UVic-UCC), Vic, Spain
Ana Dueso-Barroso
The Wellcome Trust, London, UK
Michael Dunn
The Hospital for Sick Children, Toronto, ON, Canada
Lewis Jonathan Dursi
Department of Pathology, Queen Elizabeth University Hospital, Glasgow, UK
Fraser R. Duthie
Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
Ken Dutton-Regester, Nicholas K. Hayward, Oliver Holmes, Peter A. Johansson, Stephen H. Kazakoff, Conrad R. Leonard, Felicity Newell, Katia Nones, Ann-Marie Patch, John V. Pearson, Antonia L. Pritchard, Michael C. Quinn, Paresh Vyas, Nicola Waddell, Scott Wood & Qinying Xu
Department of Oncology, Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge, UK
Douglas F. Easton
Department of Public Health and Primary Care, Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge, UK
Prostate Cancer Canada, Toronto, ON, Canada
Stuart Edmonds
Paul A. Edwards, Anthony R. Green, Andy G. Lynch, Florian Markowetz & Thomas J. Mitchell
Department of Laboratory Medicine, Translational Cancer Research, Lund University Cancer Center at Medicon Village, Lund University, Lund, Sweden
Anna Ehinger
Juergen Eils, Roland Eils & Daniel Hübschmann
New BIH Digital Health Center, Berlin Institute of Health (BIH) and Charité - Universitätsmedizin Berlin, Berlin, Germany
Juergen Eils, Roland Eils & Chris Lawerenz
CIBER Epidemiología y Salud Pública (CIBERESP), Madrid, Spain
Georgia Escaramis
Research Group on Statistics, Econometrics and Health (GRECS), UdG, Barcelona, Spain
Quantitative Genomics Laboratories (qGenomics), Barcelona, Spain
Xavier Estivill
Icelandic Cancer Registry, Icelandic Cancer Society, Reykjavik, Iceland
Jorunn E. Eyfjord, Holmfridur Hilmarsdottir & Jon G. Jonasson
State Key Laboratory of Cancer Biology, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Shaanxi, China
Daiming Fan & Yongzhan Nie
Department of Medicine (DIMED), Surgical Pathology Unit, University of Padua, Padua, Italy
Matteo Fassan
Rigshospitalet, Copenhagen, Denmark
Francesco Favero
Center for Cancer Genomics, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA
Martin L. Ferguson
Department of Biochemistry and Molecular Medicine, University of Montreal, Montreal, QC, Canada
Vincent Ferretti
Australian Institute of Tropical Health and Medicine, James Cook University, Douglas, QLD, Australia
Matthew A. Field
Department of Neuro-Oncology, Istituto Neurologico Besta, Milano, Italy
Gaetano Finocchiaro
Bioplatforms Australia, North Ryde, NSW, Australia
Anna Fitzgerald & Catherine A. Shang
Department of Pathology (Research), University College London Cancer Institute, London, UK
Adrienne M. Flanagan
Department of Surgical Oncology, Princess Margaret Cancer Centre, Toronto, ON, Canada
Neil E. Fleshner
Department of Medical Oncology, Josephine Nefkens Institute and Cancer Genomics Centre, Erasmus Medical Center, Rotterdam, CN, The Netherlands
John A. Foekens, John W. M. Martens, F. Germán Rodríguez-González, Anieta M. Sieuwerts & Marcel Smid
The University of Queensland Thoracic Research Centre, The Prince Charles Hospital, Brisbane, QLD, Australia
Kwun M. Fong
CIBIO/InBIO - Research Center in Biodiversity and Genetic Resources, Universidade do Porto, Vairão, Portugal
Nuno A. Fonseca
HCA Laboratories, London, UK
Christopher S. Foster
University of Liverpool, Liverpool, UK
The Azrieli Faculty of Medicine, Bar-Ilan University, Safed, Israel
Milana Frenkel-Morgenstern
Department of Neurosurgery, University of Florida, Gainesville, FL, USA
William Friedman
Department of Pathology, Graduate School of Medicine, University of Tokyo, Tokyo, Japan
Masashi Fukayama & Tetsuo Ushiku
University of Milano Bicocca, Monza, Italy
Carlo Gambacorti-Passerini
BGI-Shenzhen, Shenzhen, China
Shengjie Gao, Yong Hou, Chang Li, Lin Li, Siliang Li, Xiaobo Li, Xinyue Li, Dongbing Liu, Xingmin Liu, Qiang Pan-Hammarström, Hong Su, Jian Wang, Kui Wu, Heng Xiong, Huanming Yang, Chen Ye, Xiuqing Zhang, Yong Zhou & Shida Zhu
Department of Pathology, Oslo University Hospital Ulleval, Oslo, Norway
Øystein Garred
Center for Biomedical Informatics, Harvard Medical School, Boston, MA, USA
Nils Gehlenborg
Department Biochemistry and Molecular Biomedicine, University of Barcelona, Barcelona, Spain
Josep L. L. Gelpi
Office of Cancer Genomics, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA
Daniela S. Gerhard
Cancer Epigenomics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Clarissa Gerhauser, Christoph Plass & Dieter Weichenhan
Department of Cancer Biology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Jeffrey E. Gershenwald
Department of Surgical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Department of Computer Science, Yale University, New Haven, CT, USA
Mark Gerstein & Fabio C. P. Navarro
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, USA
Mark Gerstein, Sushant Kumar, Lucas Lochovsky, Shaoke Lou, Patrick D. McGillivray, Fabio C. P. Navarro, Leonidas Salichos & Jonathan Warrell
Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA
Mark Gerstein, Arif O. Harmanci, Sushant Kumar, Donghoon Lee, Shantao Li, Xiaotong Li, Lucas Lochovsky, Shaoke Lou, William Meyerson, Leonidas Salichos, Jonathan Warrell, Jing Zhang & Yan Zhang
Center for Cancer Research, Massachusetts General Hospital, Boston, MA, USA
Gad Getz & Paz Polak
Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Ronald Ghossein, Dilip D. Giri, Christine A. Iacobuzio-Donahue, Jorge Reis-Filho & Victor Reuter
Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN, USA
Nasra H. Giama, Catherine D. Moser & Lewis R. Roberts
University of Sydney, Sydney, NSW, Australia
Anthony J. Gill & James G. Kench
University of Oxford, Oxford, UK
Pelvender Gill, Freddie C. Hamdy, Katalin Karaszi, Adam Lambert, Luke Marsden, Clare Verrill & Paresh Vyas
Department of Surgery, Academic Urology Group, University of Cambridge, Cambridge, UK
Vincent J. Gnanapragasam
Department of Medicine II, University of Würzburg, Wuerzburg, Germany
Maria Elisabeth Goebler
Sylvester Comprehensive Cancer Center, University of Miami, Miami, FL, USA
Carmen Gomez
Institut Hospital del Mar d’Investigacions Mèdiques (IMIM), Barcelona, Spain
Abel Gonzalez-Perez
Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences (NIEHS), Durham, NC, USA
Dmitry A. Gordenin & Natalie Saini
St. Thomas’s Hospital, London, UK
James Gossage
Osaka International Cancer Center, Osaka, Japan
Kunihito Gotoh
Department of Pathology, Skåne University Hospital, Lund University, Lund, Sweden
Dorthe Grabau
Department of Medical Oncology, Beatson West of Scotland Cancer Centre, Glasgow, UK
Janet S. Graham
National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA
Eric Green, Carolyn M. Hutter & Heidi J. Sofia
Centre for Cancer Research, Victorian Comprehensive Cancer Centre, University of Melbourne, Melbourne, VIC, Australia
Sean M. Grimmond
Department of Medicine, Section of Hematology/Oncology, University of Chicago, Chicago, IL, USA
Robert L. Grossman
German Center for Infection Research (DZIF), Partner Site Hamburg-Borstel-Lübeck-Riems, Hamburg, Germany
Adam Grundhoff
Bioinformatics Research Centre (BiRC), Aarhus University, Aarhus, Denmark
Qianyun Guo, Asger Hobolth & Jakob Skou Pedersen
Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi, Delhi, India
Shailja Gupta & K. VijayRaghavan
National Cancer Centre Singapore, Singapore, Singapore
Jonathan Göke
Brandeis University, Waltham, MA, USA
James E. Haber
Department of Urologic Sciences, University of British Columbia, Vancouver, BC, Canada
Department of Internal Medicine, Stanford University, Stanford, CA, USA
Mark P. Hamilton
The University of Texas Health Science Center at Houston, Houston, TX, USA
Leng Han, Yang Yang & Xuanping Zhang
Imperial College NHS Trust, Imperial College, London, INY, UK
George B. Hanna
Senckenberg Institute of Pathology, University of Frankfurt Medical School, Frankfurt, Germany
Martin Hansmann
Department of Medicine, Division of Biomedical Informatics, UC San Diego School of Medicine, San Diego, CA, USA
Olivier Harismendy
Center for Precision Health, School of Biomedical Informatics, The University of Texas Health Science Center, Houston, TX, USA
Arif O. Harmanci
Oxford Nanopore Technologies, New York, NY, USA
Eoghan Harrington & Sissel Juul
Institute of Medical Science, University of Tokyo, Tokyo, Japan
Takanori Hasegawa, Shuto Hayashi, Seiya Imoto, Mitsuhiro Komura, Satoru Miyano, Naoki Miyoshi, Kazuhiro Ohi, Eigo Shimizu, Yuichi Shiraishi, Hiroko Tanaka & Rui Yamaguchi
Howard Hughes Medical Institute, University of California Santa Cruz, Santa Cruz, CA, USA
David Haussler
Wakayama Medical University, Wakayama, Japan
Shinya Hayami, Masaki Ueno & Hiroki Yamaue
Department of Internal Medicine, Division of Medical Oncology, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
D. Neil Hayes
University of Tennessee Health Science Center for Cancer Research, Memphis, TN, USA
Department of Histopathology, Salford Royal NHS Foundation Trust, Salford, UK
Stephen J. Hayes
Faculty of Biology, Medicine and Health, University of Manchester, Manchester, UK
BIOPIC, ICG and College of Life Sciences, Peking University, Beijing, China
Yao He & Zemin Zhang
Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, China
Children’s Hospital of Philadelphia, Philadelphia, PA, USA
Allison P. Heath
Department of Bioinformatics and Computational Biology and Department of Systems Biology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Apurva M. Hegde, Yiling Lu & John N. Weinstein
Karolinska Institute, Stockholm, Sweden
Eva Hellstrom-Lindberg & Jesper Lagergren
The Donnelly Centre, University of Toronto, Toronto, ON, Canada
Mohamed Helmy & Jeffrey A. Wintersinger
Department of Medical Genetics, College of Medicine, Hallym University, Chuncheon, South Korea
Seong Gu Heo, Eun Pyo Hong & Ji Wan Park
Department of Experimental and Health Sciences, Institute of Evolutionary Biology (UPF-CSIC), Universitat Pompeu Fabra, Barcelona, Spain
José María Heredia-Genestar, Tomas Marques-Bonet & Arcadi Navarro
Health Data Science Unit, University Clinics, Heidelberg, Germany
Carl Herrmann
Massachusetts General Hospital Center for Cancer Research, Charlestown, MA, USA
Julian M. Hess & Yosef E. Maruvka
Hokkaido University, Sapporo, Japan
Satoshi Hirano & Toru Nakamura
Department of Pathology and Clinical Laboratory, National Cancer Center Hospital, Tokyo, Japan
Nobuyoshi Hiraoka
Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Katherine A. Hoadley & Tara J. Skelly
Computational Biology, Leibniz Institute on Aging - Fritz Lipmann Institute (FLI), Jena, Germany
Steve Hoffmann
University of Melbourne Centre for Cancer Research, Melbourne, VIC, Australia
Oliver Hofmann
University of Nebraska Medical Center, Omaha, NE, USA
Michael A. Hollingsworth & Sarah P. Thayer
Syntekabio Inc, Daejeon, South Korea
Jongwhi H. Hong
Department of Pathology, Academic Medical Center, Amsterdam, AZ, The Netherlands
Gerrit K. Hooijer
China National GeneBank-Shenzhen, Shenzhen, China
Yong Hou, Chang Li, Siliang Li, Xiaobo Li, Dongbing Liu, Xingmin Liu, Henk G. Stunnenberg, Hong Su, Kui Wu, Heng Xiong, Chen Ye & Shida Zhu
Division of Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Volker Hovestadt, Murat Iskar, Peter Lichter, Bernhard Radlwimmer & Marc Zapatka
Division of Life Science and Applied Genomics Center, Hong Kong University of Science and Technology, Clear Water Bay, Hong Kong, China
Taobo Hu, Yogesh Kumar, Eric Z. Ma, Zhenggang Wu & Hong Xue
Icahn School of Medicine at Mount Sinai, New York, NY, USA
Kuan-lin Huang
Geneplus-Shenzhen, Shenzhen, China
School of Computer Science and Technology, Xi’an Jiaotong University, Xi’an, China
Yi Huang, Jiayin Wang, Xiao Xiao & Xuanping Zhang
AbbVie, North Chicago, IL, USA
Thomas J. Hudson
Institute of Pathology, Charité – University Medicine Berlin, Berlin, Germany
Michael Hummel & Dido Lenze
Centre for Translational and Applied Genomics, British Columbia Cancer Agency, Vancouver, BC, Canada
David Huntsman
Edinburgh Royal Infirmary, Edinburgh, UK
Ted R. Hupp
Berlin Institute for Medical Systems Biology, Max Delbrück Center for Molecular Medicine, Berlin, Germany
Matthew R. Huska, Julia Markowski & Roland F. Schwarz
Department of Pediatric Immunology, Hematology and Oncology, University Hospital, Heidelberg, Germany
Daniel Hübschmann
Daniel Hübschmann, Christof von Kalle & Roland F. Schwarz
Heidelberg Institute for Stem Cell Technology and Experimental Medicine (HI-STEM), Heidelberg, Germany
Institute for Computational Biomedicine, Weill Cornell Medical College, New York, NY, USA
Marcin Imielinski
Marcin Imielinski & Xiaotong Yao
Department of Urology, James Buchanan Brady Urological Institute, Johns Hopkins University School of Medicine, Baltimore, MD, USA
William B. Isaacs
Department of Preventive Medicine, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
Shumpei Ishikawa, Hiroto Katoh & Daisuke Komura
Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX, USA
Michael Ittmann
Department of Pathology and Immunology, Baylor College of Medicine, Houston, TX, USA
Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA
Technical University of Denmark, Lyngby, Denmark
Jose M. G. Izarzugaza
Department of Pathology, College of Medicine, Hanyang University, Seoul, South Korea
Jocelyne Jacquemier, Hyung-Yong Kim & Gu Kong
Academic Unit of Surgery, School of Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow Royal Infirmary, Glasgow, UK
Nigel B. Jamieson
Department of Pathology, Asan Medical Center, College of Medicine, Ulsan University, Songpa-gu, Seoul, South Korea
Se Jin Jang & Hee Jin Lee
Science Writer, Garrett Park, MD, USA
Karine Jegalian
International Cancer Genome Consortium (ICGC)/ICGC Accelerating Research in Genomic Oncology (ARGO) Secretariat, Ontario Institute for Cancer Research, Toronto, ON, Canada
Jennifer L. Jennings
Lara Jerman
Department of Public Health Sciences, University of Chicago, Chicago, IL, USA
Research Institute, NorthShore University HealthSystem, Evanston, IL, USA
Department for Biomedical Research, University of Bern, Bern, Switzerland
Rory Johnson, Andrés Lanzós & Mark A. Rubin
Centre of Genomics and Policy, McGill University and Génome Québec Innovation Centre, Montreal, QC, Canada
Yann Joly, Bartha M. Knoppers, Mark Phillips & Adrian Thorogood
Carolina Center for Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Corbin D. Jones
Hopp Children’s Cancer Center (KiTZ), Heidelberg, Germany
David T. W. Jones, Marcel Kool & Stefan M. Pfister
Pediatric Glioma Research Group, German Cancer Research Center (DKFZ), Heidelberg, Germany
David T. W. Jones
Cancer Research UK, London, UK
Nic Jones & David Scott
Indivumed GmbH, Hamburg, Germany
Hartmut Juhl
Genome Integration Data Center, Syntekabio, Inc, Daejeon, South Korea
Jongsun Jung
University Hospital Zurich, Zurich, Switzerland
Andre Kahles, Kjong-Van Lehmann & Gunnar Rätsch
Clinical Bioinformatics, Swiss Institute of Bioinformatics, Geneva, Switzerland
Abdullah Kahraman
Institute for Pathology and Molecular Pathology, University Hospital Zurich, Zurich, Switzerland
Institute of Molecular Life Sciences, University of Zurich, Zurich, Switzerland
Abdullah Kahraman & Christian von Mering
MRC Human Genetics Unit, MRC IGMM, University of Edinburgh, Edinburgh, UK
Vera B. Kaiser & Colin A. Semple
Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA
Beth Karlan
Department of Biology, Bioinformatics Group, Division of Molecular Biology, Faculty of Science, University of Zagreb, Zagreb, Croatia
Rosa Karlić
Department for Internal Medicine II, University Hospital Schleswig-Holstein, Kiel, Germany
Dennis Karsch & Michael Kneba
Genetics and Molecular Pathology, SA Pathology, Adelaide, SA, Australia
Karin S. Kassahn
Department of Gastric Surgery, National Cancer Center Hospital, Tokyo, Japan
Hitoshi Katai
Department of Bioinformatics, Division of Cancer Genomics, National Cancer Center Research Institute, Tokyo, Japan
Mamoru Kato, Hirofumi Rokutan & Mihoko Saito-Adachi
A.A. Kharkevich Institute of Information Transmission Problems, Moscow, Russia
Marat D. Kazanov
Oncology and Immunology, Dmitry Rogachev National Research Center of Pediatric Hematology, Moscow, Russia
Skolkovo Institute of Science and Technology, Moscow, Russia
Department of Surgery, The George Washington University, School of Medicine and Health Science, Washington, DC, USA
Electron Kebebew
Endocrine Oncology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA
Melanoma Institute Australia, Macquarie University, Sydney, NSW, Australia
Richard F. Kefford
MIT Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, USA
Manolis Kellis
Tissue Pathology and Diagnostic Oncology, Royal Prince Alfred Hospital, Sydney, NSW, Australia
James G. Kench & Richard A. Scolyer
Cholangiocarcinoma Screening and Care Program and Liver Fluke and Cholangiocarcinoma Research Centre, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand
Narong Khuntikeo
Controlled Department and Institution, New York, NY, USA
Ekta Khurana
Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY, USA
Ekta Khurana & Alexander Martinez-Fundichely
National Cancer Center, Gyeonggi, South Korea
Hark Kyun Kim
Department of Biochemistry, College of Medicine, Ewha Womans University, Seoul, South Korea
Hyung-Lae Kim
Health Sciences Department of Biomedical Informatics, University of California San Diego, La Jolla, CA, USA
Research Core Center, National Cancer Centre Korea, Goyang-si, South Korea
Jong K. Kim
Department of Health Sciences and Technology, Sungkyunkwan University School of Medicine, Seoul, South Korea
Youngwook Kim
Samsung Genome Institute, Seoul, South Korea
Breast Oncology Program, Dana-Farber/Brigham and Women’s Cancer Center, Boston, MA, USA
Tari A. King
Department of Surgery, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Tari A. King & Samuel Singer
Division of Breast Surgery, Brigham and Women’s Hospital, Boston, MA, USA
Integrative Bioinformatics Support Group, National Institute of Environmental Health Sciences (NIEHS), Durham, NC, USA
Leszek J. Klimczak
Department of Clinical Science, University of Bergen, Bergen, Norway
Stian Knappskog & Ola Myklebost
Center For Medical Innovation, Seoul National University Hospital, Seoul, South Korea
Youngil Koh
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
Youngil Koh & Sung-Soo Yoon
Institute of Computer Science, Polish Academy of Sciences, Warsawa, Poland
Jan Komorowski
Functional and Structural Genomics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Marcel Kool, Andrey Korshunov, Michael Koscher, Stefan M. Pfister & Qi Wang
Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, , National Institutes of Health, Bethesda, MD, USA
Roelof Koster
Institute for Medical Informatics Statistics and Epidemiology, University of Leipzig, Leipzig, Germany
Markus Kreuz & Markus Loeffler
Morgan Welch Inflammatory Breast Cancer Research Program and Clinic, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Savitri Krishnamurthy
Department of Hematology and Oncology, Georg-Augusts-University of Göttingen, Göttingen, Germany
Dieter Kube & Lorenz H. P. Trümper
Institute of Cell Biology (Cancer Research), University of Duisburg-Essen, Essen, Germany
Ralf Küppers
King’s College London and Guy’s and St. Thomas’ NHS Foundation Trust, London, UK
Jesper Lagergren
Center for Epigenetics, Van Andel Research Institute, Grand Rapids, MI, USA
Peter W. Laird
The University of Queensland Centre for Clinical Research, Royal Brisbane and Women’s Hospital, Herston, QLD, Australia
Sunil R. Lakhani & Peter T. Simpson
Department of Pediatric Oncology and Hematology, University of Cologne, Cologne, Germany
Pablo Landgraf
University of Düsseldorf, Düsseldorf, Germany
Pablo Landgraf & Guido Reifenberger
Department of Pathology, Institut Jules Bordet, Brussels, Belgium
Denis Larsimont
Institute of Biomedicine, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden
Erik Larsson
Children’s Medical Research Institute, Sydney, NSW, Australia
Loretta M. S. Lau & Hilda A. Pickett
ILSbio, LLC Biobank, Chestertown, MD, USA
Division of Genetics and Genomics, Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA
Eunjung Alice Lee
Institute for Bioengineering and Biopharmaceutical Research (IBBR), Hanyang University, Seoul, South Korea
Jeong-Yeon Lee
Department of Statistics, University of California Santa Cruz, Santa Cruz, CA, USA
National Genotyping Center, Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan
Ming Ta Michael Lee
Department of Vertebrate Genomics/Otto Warburg Laboratory Gene Regulation and Systems Biology of Cancer, Max Planck Institute for Molecular Genetics, Berlin, Germany
Hans Lehrach, Hans-Jörg Warnatz & Marie-Laure Yaspo
McGill University and Genome Quebec Innovation Centre, Montreal, QC, Canada
Louis Letourneau
biobyte solutions GmbH, Heidelberg, Germany
Ivica Letunic
Gynecologic Oncology, NYU Laura and Isaac Perlmutter Cancer Center, New York University, New York, NY, USA
Douglas A. Levine
Division of Oncology, Stem Cell Biology Section, Washington University School of Medicine, St. Louis, MO, USA
Department of Systems Biology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Harvard University, Cambridge, MA, USA
Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA
W. M. Linehan
University of Oslo, Oslo, Norway
Ole Christian Lingjærde & Torill Sauer
University of Toronto, Toronto, ON, Canada
Fei-Fei Fei Liu, Quaid D. Morris, Ruian Shi, Shankar Vembu & Fan Yang
Peking University, Beijing, China
Fenglin Liu, Fan Zhang, Liangtao Zheng & Xiuqing Zheng
School of Life Sciences, Peking University, Beijing, China
Fenglin Liu
Leidos Biomedical Research, Inc, McLean, VA, USA
Hematology, Hospital Clinic, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), University of Barcelona, Barcelona, Spain
Armando Lopez-Guillermo
Second Military Medical University, Shanghai, China
Yong-Jie Lu & Hongwei Zhang
Chinese Cancer Genome Consortium, Shenzhen, China
Department of Medical Oncology, Beijing Hospital, Beijing, China
Laboratory of Molecular Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital and Institute, Beijing, China
Youyong Lu & Rui Xing
School of Medicine/School of Mathematics and Statistics, University of St. Andrews, St, Andrews, Fife, UK
Andy G. Lynch
Institute for Systems Biology, Seattle, WA, USA
Lisa Lype, Sheila M. Reynolds & Ilya Shmulevich
Department of Biochemistry and Molecular Biology, Faculty of Medicine, University Institute of Oncology-IUOPA, Oviedo, Spain
Carlos López-Otín & Xose S. Puente
Institut Bergonié, Bordeaux, France
Gaetan MacGrogan
Cancer Unit, MRC University of Cambridge, Cambridge, UK
Shona MacRae
Department of Pathology and Laboratory Medicine, Center for Personalized Medicine, Children’s Hospital Los Angeles, Los Angeles, CA, USA
Dennis T. Maglinte
John Curtin School of Medical Research, Canberra, ACT, Australia
Graham J. Mann
MVZ Department of Oncology, PraxisClinic am Johannisplatz, Leipzig, Germany
Luisa Mantovani-Löffler
Department of Information Technology, Ghent University, Ghent, Belgium
Kathleen Marchal & Sergio Pulido-Tamayo
Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
Kathleen Marchal, Sergio Pulido-Tamayo & Lieven P. C. Verbeke
Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH, USA
Elaine R. Mardis
Computational Biology Program, School of Medicine, Oregon Health and Science University, Portland, OR, USA
Adam A. Margolin & Adam J. Struck
Department of Surgery, Duke University, Durham, NC, USA
Jeffrey Marks
Institució Catalana de Recerca i Estudis Avançats (ICREA), Barcelona, Spain
Tomas Marques-Bonet, Jose I. Martin-Subero, Arcadi Navarro, David Torrents & Alfonso Valencia
Institut Català de Paleontologia Miquel Crusafont, Universitat Autònoma de Barcelona, Barcelona, Spain
Tomas Marques-Bonet
Sancha Martin & Ke Yuan
Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain
Jose I. Martin-Subero
Division of Oncology, Washington University School of Medicine, St. Louis, MO, USA
R. Jay Mashl
Department of Surgery and Cancer, Imperial College, London, INY, UK
Applications Department, Oxford Nanopore Technologies, Oxford, UK
Simon Mayes & Daniel J. Turner
Department of Obstetrics, Gynecology and Reproductive Services, University of California San Francisco, San Francisco, CA, USA
Karen McCune & Karen Smith-McCune
Department of Biochemistry and Molecular Medicine, University California at Davis, Sacramento, CA, USA
John D. McPherson
STTARR Innovation Facility, Princess Margaret Cancer Centre, Toronto, ON, Canada
Discipline of Surgery, Western Sydney University, Penrith, NSW, Australia
Neil D. Merrett
Yale School of Medicine, Yale University, New Haven, CT, USA
William Meyerson
Department of Genetics, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Piotr A. Mieczkowski, Joel S. Parker, Charles M. Perou, Donghui Tan, Umadevi Veluvolu & Matthew D. Wilkerson
Departments of Neurology and Neurosurgery, Henry Ford Hospital, Detroit, MI, USA
Tom Mikkelsen
Precision Oncology, OHSU Knight Cancer Institute, Oregon Health and Science University, Portland, OR, USA
Gordon B. Mills
Institute of Pathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
Sarah Minner, Guido Sauter & Ronald Simon
Department of Health Sciences, Faculty of Medical Sciences, Kyushu University, Fukuoka, Japan
Shinichi Mizuno
Heidelberg Academy of Sciences and Humanities, Heidelberg, Germany
Fruzsina Molnár-Gábor
Department of Clinical Pathology, University of Melbourne, Melbourne, VIC, Australia
Carl Morrison, Karin A. Oien, Chawalit Pairojkul, Paul M. Waring & Marc J. van de Vijver
Department of Pathology, Roswell Park Cancer Institute, Buffalo, NY, USA
Carl Morrison
Department of Computer Science, University of Helsinki, Helsinki, Finland
Ville Mustonen
Institute of Biotechnology, University of Helsinki, Helsinki, Finland
Organismal and Evolutionary Biology Research Programme, University of Helsinki, Helsinki, Finland
Department of Obstetrics and Gynecology, Division of Gynecologic Oncology, Washington University School of Medicine, St. Louis, MO, USA
David Mutch
Penrose St. Francis Health Services, Colorado Springs, CO, USA
Jerome Myers
Institute of Pathology, Ulm University and University Hospital of Ulm, Ulm, Germany
Peter Möller
National Cancer Center, Tokyo, Japan
Hitoshi Nakagama
Genome Institute of Singapore, Singapore, Singapore
Tannistha Nandi & Patrick Tan
32Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA
Fabio C. P. Navarro
German Cancer Aid, Bonn, Germany
Gerd Nettekoven & Laura Planko
Programme in Cancer and Stem Cell Biology, Centre for Computational Biology, Duke-NUS Medical School, Singapore, Singapore
Alvin Wei Tian Ng
The Chinese University of Hong Kong, Shatin, NT, Hong Kong, China
Fourth Military Medical University, Shaanxi, China
Yongzhan Nie
The University of Cambridge School of Clinical Medicine, Cambridge, UK
Serena Nik-Zainal
St. Jude Children’s Research Hospital, Memphis, TN, USA
Paul A. Northcott
University Health Network, Princess Margaret Cancer Centre, Toronto, ON, Canada
Faiyaz Notta & Ming Tsao
Center for Biomolecular Science and Engineering, University of California Santa Cruz, Santa Cruz, CA, USA
Brian D. O’Connor
Department of Medicine, University of Chicago, Chicago, IL, USA
Peter O’Donnell
Department of Neurology, Mayo Clinic, Rochester, MN, USA
Brian Patrick O’Neill
Cambridge Oesophagogastric Centre, Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK
J. Robert O’Neill
Institute of Cancer Sciences, College of Medical Veterinary and Life Sciences, University of Glasgow, Glasgow, UK
Karin A. Oien
Department of Epidemiology, University of Alabama at Birmingham, Birmingham, AL, USA
Akinyemi I. Ojesina
HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA
O’Neal Comprehensive Cancer Center, University of Alabama at Birmingham, Birmingham, AL, USA
Department of Pathology, Keio University School of Medicine, Tokyo, Japan
Hidenori Ojima
Department of Hepatobiliary and Pancreatic Oncology, National Cancer Center Hospital, Tokyo, Japan
Takuji Okusaka
Sage Bionetworks, Seattle, WA, USA
Larsson Omberg
Lymphoma Genomic Translational Research Laboratory, National Cancer Centre, Singapore, Singapore
Choon Kiat Ong
Department of Clinical Pathology, Robert-Bosch-Hospital, Stuttgart, Germany
Department of Cell and Systems Biology, University of Toronto, Toronto, ON, Canada
B. F. Francis Ouellette
Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden
Qiang Pan-Hammarström
Center for Liver Cancer, Research Institute and Hospital, National Cancer Center, Gyeonggi, South Korea
Joong-Won Park
Division of Hematology-Oncology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, South Korea
Keunchil Park
Samsung Advanced Institute for Health Sciences and Technology, Sungkyunkwan University School of Medicine, Seoul, South Korea
Cheonan Industry-Academic Collaboration Foundation, Sangmyung University, Cheonan, South Korea
Kiejung Park
NYU Langone Medical Center, New York, NY, USA
Harvey Pass
Department of Hematology and Medical Oncology, Cleveland Clinic, Cleveland, OH, USA
Nathan A. Pennell
Department of Radiation Oncology, University of California San Francisco, San Francisco, CA, USA
Marc D. Perry
Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA
Gloria M. Petersen
Helen F. Graham Cancer Center at Christiana Care Health Systems, Newark, DE, USA
Nicholas Petrelli
Heidelberg University Hospital, Heidelberg, Germany
Stefan M. Pfister
CSRA Incorporated, Fairfax, VA, USA
Todd D. Pihl
Research Department of Pathology, University College London Cancer Institute, London, UK
Nischalan Pillay
Department of Research Oncology, Guy’s Hospital, King’s Health Partners AHSC, King’s College London School of Medicine, London, UK
Sarah Pinder
Faculty of Medicine and Health Sciences, Macquarie University, Sydney, NSW, Australia
Andreia V. Pinho
University Hospital of Minjoz, INSERM UMR 1098, Besançon, France
Xavier Pivot
Spanish National Cancer Research Centre, Madrid, Spain
Center of Digestive Diseases and Liver Transplantation, Fundeni Clinical Institute, Bucharest, Romania
Irinel Popescu
Cureline, Inc, South San Francisco, CA, USA
Olga Potapova
St. Luke’s Cancer Centre, Royal Surrey County Hospital NHS Foundation Trust, Guildford, UK
Shaun R. Preston
Cambridge Breast Unit, Addenbrooke’s Hospital, Cambridge University Hospital NHS Foundation Trust and NIHR Cambridge Biomedical Research Centre, Cambridge, UK
Elena Provenzano
East of Scotland Breast Service, Ninewells Hospital, Aberdeen, UK
Colin A. Purdie
Department of Genetics, Microbiology and Statistics, University of Barcelona, IRSJD, IBUB, Barcelona, Spain
Raquel Rabionet
Department of Obstetrics and Gynecology, Medical College of Wisconsin, Milwaukee, WI, USA
Janet S. Rader
Hematology and Medical Oncology, Winship Cancer Institute of Emory University, Atlanta, GA, USA
Suresh Ramalingam
Benjamin J. Raphael & Matthew A. Reyna
Vanderbilt Ingram Cancer Center, Vanderbilt University, Nashville, TN, USA
W. Kimryn Rathmell
Ohio State University College of Medicine and Arthur G. James Comprehensive Cancer Center, Columbus, OH, USA
Matthew Ringel
Department of Surgery, Yokohama City University Graduate School of Medicine, Kanagawa, Japan
Yasushi Rino
Division of Chromatin Networks, German Cancer Research Center (DKFZ) and BioQuant, Heidelberg, Germany
Karsten Rippe
Research Computing Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Jeffrey Roach
School of Molecular Biosciences and Center for Reproductive Biology, Washington State University, Pullman, WA, USA
Steven A. Roberts
Finsen Laboratory and Biotech Research and Innovation Centre (BRIC), University of Copenhagen, Copenhagen, Denmark
F. Germán Rodríguez-González, Nikos Sidiropoulos & Joachim Weischenfeldt
Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada
Michael H. A. Roehrl & Stefano Serra
Department of Pathology, Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Michael H. A. Roehrl
University Hospital Giessen, Pediatric Hematology and Oncology, Giessen, Germany
Marius Rohde
Oncologie Sénologie, ICM Institut Régional du Cancer, Montpellier, France
Gilles Romieu
Institute of Clinical Molecular Biology, Christian-Albrechts-University, Kiel, Germany
Philip C. Rosenstiel & Markus B. Schilhabel
Institute of Pathology, University of Wuerzburg, Wuerzburg, Germany
Andreas Rosenwald
Department of Urology, North Bristol NHS Trust, Bristol, UK
Edward W. Rowe
SingHealth, Duke-NUS Institute of Precision Medicine, National Heart Centre Singapore, Singapore, Singapore
Steven G. Rozen, Patrick Tan & Bin Tean Teh
Department of Computer Science, University of Toronto, Toronto, ON, Canada
Yulia Rubanova, Jared T. Simpson & Jeffrey A. Wintersinger
Bern Center for Precision Medicine, University Hospital of Bern, University of Bern, Bern, Switzerland
Mark A. Rubin
Englander Institute for Precision Medicine, Weill Cornell Medicine and New York Presbyterian Hospital, New York, NY, USA
Meyer Cancer Center, Weill Cornell Medicine, New York, NY, USA
Pathology and Laboratory, Weill Cornell Medical College, New York, NY, USA
Vall d’Hebron Institute of Oncology: VHIO, Barcelona, Spain
Carlota Rubio-Perez
General and Hepatobiliary-Biliary Surgery, Pancreas Institute, University and Hospital Trust of Verona, Verona, Italy
Andrea Ruzzenente
National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bangalore, India
Radhakrishnan Sabarinathan
S. Cenk Sahinalp
Department of Pathology, GZA-ZNA Hospitals, Antwerp, Belgium
Roberto Salgado
Analytical Biological Services, Inc, Wilmington, DE, USA
Charles Saller
Sydney Medical School, University of Sydney, Sydney, NSW, Australia
Jaswinder S. Samra & Richard A. Scolyer
cBio Center, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA
Chris Sander & Ciyue Shen
Department of Cell Biology, Harvard Medical School, Boston, MA, USA
Advanced Centre for Treatment Research and Education in Cancer, Tata Memorial Centre, Navi Mumbai, Maharashtra, India
Rajiv Sarin
School of Environmental and Life Sciences, Faculty of Science, The University of Newcastle, Ourimbah, NSW, Australia
Christopher J. Scarlett
Department of Dermatology, University Hospital of Essen, Essen, Germany
Dirk Schadendorf
Bioinformatics and Omics Data Analytics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Matthias Schlesner
Department of Urology, Charité Universitätsmedizin Berlin, Berlin, Germany
Thorsten Schlomm & Joachim Weischenfeldt
Martini-Clinic, Prostate Cancer Center, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
Thorsten Schlomm
Department of General Internal Medicine, University of Kiel, Kiel, Germany
Stefan Schreiber
German Cancer Consortium (DKTK), Partner site Berlin, Berlin, Germany
Roland F. Schwarz
Cancer Research Institute, Beth Israel Deaconess Medical Center, Boston, MA, USA
Ralph Scully
University of Pittsburgh, Pittsburgh, PA, USA
Raja Seethala
Department of Ophthalmology and Ocular Genomics Institute, Massachusetts Eye and Ear, Harvard Medical School, Boston, MA, USA
Ayellet V. Segre
Center for Psychiatric Genetics, NorthShore University HealthSystem, Evanston, IL, USA
Subhajit Sengupta
Van Andel Research Institute, Grand Rapids, MI, USA
Hui Shen & Wanding Zhou
Laboratory of Molecular Medicine, Human Genome Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan
Tatsuhiro Shibata, Hirokazu Taniguchi & Tomoko Urushidate
Japan Agency for Medical Research and Development, Tokyo, Japan
Kiyo Shimizu & Takashi Yugawa
Seung Jun Shin & Stefan G. Stark
Murtha Cancer Center, Walter Reed National Military Medical Center, Bethesda, MD, USA
Craig Shriver
Human Genetics, University of Kiel, Kiel, Germany
Reiner Siebert
Department of Oncologic Pathology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA
Sabina Signoretti
Oregon Health and Science University, Portland, OR, USA
Jaclyn Smith
Center for RNA Interference and Noncoding RNA, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Anil K. Sood
Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Department of Gynecologic Oncology and Reproductive Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
University Hospitals Coventry and Warwickshire NHS Trust, Coventry, UK
Sharmila Sothi
Department of Radiation Oncology, Radboud University Nijmegen Medical Centre, Nijmegen, GA, The Netherlands
Paul N. Span
Institute for Genomics and Systems Biology, University of Chicago, Chicago, IL, USA
Jonathan Spring
Clinic for Hematology and Oncology, St.-Antonius-Hospital, Eschweiler, Germany
Peter Staib
Computational and Systems Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Stefan G. Stark
University of Iceland, Reykjavik, Iceland
Ólafur Andri Stefánsson
Division of Computational Genomics and Systems Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Oliver Stegle
Dundee Cancer Centre, Ninewells Hospital, Dundee, UK
Alasdair Stenhouse & Alastair M. Thompson
Department for Internal Medicine III, University of Ulm and University Hospital of Ulm, Ulm, Germany
Stephan Stilgenbauer
Institut Curie, INSERM Unit 830, Paris, France
Henk G. Stunnenberg & Anne Vincent-Salomon
Department of Gastroenterology and Hepatology, Yokohama City University Graduate School of Medicine, Kanagawa, Japan
Akihiro Suzuki
Department of Laboratory Medicine, Radboud University Nijmegen Medical Centre, Nijmegen, GA, The Netherlands
Division of Cancer Genome Research, German Cancer Research Center (DKFZ), Heidelberg, Germany
Holger Sültmann
Department of General Surgery, Singapore General Hospital, Singapore, Singapore
Benita Kiat Tee Tan
Cancer Science Institute of Singapore, National University of Singapore, Singapore, Singapore
Patrick Tan & Bin Tean Teh
Department of Medical and Clinical Genetics, Genome-Scale Biology Research Program, University of Helsinki, Helsinki, Finland
Tomas J. Tanskanen
East Anglian Medical Genetics Service, Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK
Patrick Tarpey
Irving Institute for Cancer Dynamics, Columbia University, New York, NY, USA
Simon Tavaré
Institute of Molecular and Cell Biology, Singapore, Singapore
Bin Tean Teh
Laboratory of Cancer Epigenome, Division of Medical Science, National Cancer Centre Singapore, Singapore, Singapore
Universite Lyon, INCa-Synergie, Centre Léon Bérard, Lyon, France
Gilles Thomas
Department of Urology, Mayo Clinic, Rochester, MN, USA
R. Houston Thompson
Royal National Orthopaedic Hospital - Stanmore, Stanmore, Middlesex, UK
Roberto Tirabosco
Department of Biochemistry, Genetics and Immunology, University of Vigo, Vigo, Spain
Giovanni Paolo II / I.R.C.C.S. Cancer Institute, Bari, BA, Italy
Stefania Tommasi
Neuroblastoma Genomics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Umut H. Toprak
Fondazione Policlinico Universitario Gemelli IRCCS, Rome, Italy, Rome, Italy
Giampaolo Tortora
University of Verona, Verona, Italy
Centre National de Génotypage, CEA - Institute de Génomique, Evry, France
CAPHRI Research School, Maastricht University, Maastricht, ER, The Netherlands
David Townend
Department of Biopathology, Centre Léon Bérard, Lyon, France
Isabelle Treilleux
Université Claude Bernard Lyon 1, Villeurbanne, France
Core Research for Evolutional Science and Technology (CREST), JST, Tokyo, Japan
Tatsuhiko Tsunoda
Department of Biological Sciences, Laboratory for Medical Science Mathematics, Graduate School of Science, University of Tokyo, Yokohama, Japan
Department of Medical Science Mathematics, Medical Research Institute, Tokyo Medical and Dental University (TMDU), Tokyo, Japan
Cancer Ageing and Somatic Mutation Programme, Wellcome Sanger Institute, Hinxton, UK
Jose M. C. Tubio
University Hospitals Birmingham NHS Foundation Trust, Birmingham, UK
Olga Tucker
Centre for Cancer Research and Cell Biology, Queen’s University, Belfast, UK
Richard Turkington
Breast Medical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Naoto T. Ueno
Department of Surgery, Johns Hopkins University School of Medicine, Baltimore, MD, USA
Christopher Umbricht
Department of Oncology-Pathology, Science for Life Laboratory, Karolinska Institute, Stockholm, Sweden
Husen M. Umer
School of Cancer Sciences, Faculty of Medicine, University of Southampton, Southampton, UK
Timothy J. Underwood
Department of Gene Technology, Tallinn University of Technology, Tallinn, Estonia
Liis Uusküla-Reimand
Genetics and Genome Biology Program, SickKids Research Institute, The Hospital for Sick Children, Toronto, ON, Canada
Departments of Neurosurgery and Hematology and Medical Oncology, Winship Cancer Institute and School of Medicine, Emory University, Atlanta, GA, USA
Erwin G. Van Meir
Department of Clinical and Molecular Medicine, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim, Norway
Miguel Vazquez
Argmix Consulting, North Vancouver, BC, Canada
Shankar Vembu
Department of Information Technology, Ghent University, Interuniversitair Micro-Electronica Centrum (IMEC), Ghent, Belgium
Lieven P. C. Verbeke
Nuffield Department of Surgical Sciences, John Radcliffe Hospital, University of Oxford, Oxford, UK
Clare Verrill
Institute of Mathematics and Computer Science, University of Latvia, Riga, LV, Latvia
Juris Viksna
Discipline of Pathology, Sydney Medical School, University of Sydney, Sydney, NSW, Australia
Ricardo E. Vilain
Department of Applied Mathematics and Theoretical Physics, Centre for Mathematical Sciences, University of Cambridge, Cambridge, UK
Ignacio Vázquez-García
Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Ignacio Vázquez-García & Venkata D. Yellapantula
Department of Statistics, Columbia University, New York, NY, USA
Department of Immunology, Genetics and Pathology, Science for Life Laboratory, Uppsala University, Uppsala, Sweden
Claes Wadelius
School of Electronic and Information Engineering, Xi’an Jiaotong University, Xi’an, China
Jiayin Wang & Kai Ye
Department of Histopathology, Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK
Anne Y. Warren
Oxford NIHR Biomedical Research Centre, University of Oxford, Oxford, UK
David C. Wedge
Georgia Regents University Cancer Center, Augusta, GA, USA
Paul Weinberger
Wythenshawe Hospital, Manchester, UK
Department of Genetics, Washington University School of Medicine, St.Louis, MO, USA
Michael C. Wendl
Department of Biological Oceanography, Leibniz Institute of Baltic Sea Research, Rostock, Germany
Johannes Werner
Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
Justin P. Whalley
Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA
Thoracic Oncology Laboratory, Mayo Clinic, Rochester, MN, USA
Dennis Wigle
Richard K. Wilson
Department of Obstetrics and Gynecology, Division of Gynecologic Oncology, Mayo Clinic, Rochester, MN, USA
Boris Winterhoff
International Institute for Molecular Oncology, Poznań, Poland
Maciej Wiznerowicz
Poznan University of Medical Sciences, Poznań, Poland
Genomics and Proteomics Core Facility High Throughput Sequencing Unit, German Cancer Research Center (DKFZ), Heidelberg, Germany
Stephan Wolf
NCCS-VARI Translational Research Laboratory, National Cancer Centre Singapore, Singapore, Singapore
Bernice H. Wong
Edison Family Center for Genome Sciences and Systems Biology, Washington University, St. Louis, MO, USA
Winghing Wong
MRC-University of Glasgow Centre for Virus Research, Glasgow, UK
Derek W. Wright
Department of Medical Informatics and Clinical Epidemiology, Division of Bioinformatics and Computational Biology, OHSU Knight Cancer Institute, Oregon Health and Science University, Portland, OR, USA
Guanming Wu
School of Electronic Information and Communications, Huazhong University of Science and Technology, Wuhan, China
Department of Applied Mathematics and Statistics, Johns Hopkins University, Baltimore, MD, USA
Department of Cancer Genome Informatics, Graduate School of Medicine, Osaka University, Osaka, Japan
Shinichi Yachida
Institute of Computer Science, Heidelberg University, Heidelberg, Germany
Sergei Yakneen
School of Mathematics and Statistics, University of Sydney, Sydney, NSW, Australia
Jean Y. Yang
Ben May Department for Cancer Research, University of Chicago, Chicago, IL, USA
Lixing Yang
Department of Human Genetics, University of Chicago, Chicago, IL, USA
Tri-Institutional PhD Program in Computational Biology and Medicine, Weill Cornell Medicine, New York, NY, USA
Xiaotong Yao
The First Affiliated Hospital, Xi’an Jiaotong University, Xi’an, China
Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Shatin, NT, Hong Kong, China
Department of Biostatistics, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Kaixian Yu & Hongtu Zhu
Duke-NUS Medical School, Singapore, Singapore
Department of Surgery, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
School of Computing Science, University of Glasgow, Glasgow, UK
Division of Orthopaedic Surgery, Oslo University Hospital, Oslo, Norway
Olga Zaikova
Eastern Clinical School, Monash University, Melbourne, VIC, Australia
Nikolajs Zeps
Epworth HealthCare, Richmond, VIC, Australia
Department of Biostatistics and Computational Biology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA
Cheng-Zhong Zhang
Department of Biomedical Informatics, College of Medicine, The Ohio State University, Columbus, OH, USA
The Ohio State University Comprehensive Cancer Center (OSUCCC – James), Columbus, OH, USA
The University of Texas School of Biomedical Informatics (SBMI) at Houston, Houston, TX, USA
Zhongming Zhao
Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Department of Biochemistry and Molecular Genetics, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA
Faculty of Medicine and Health, University of Sydney, Sydney, NSW, Australia
Anna deFazio
Department of Pathology, Erasmus Medical Center Rotterdam, Rotterdam, GD, The Netherlands
Carolien H. M. van Deurzen
Division of Molecular Carcinogenesis, The Netherlands Cancer Institute, Amsterdam, CX, The Netherlands
L. van’t Veer
Institute of Molecular Life Sciences and Swiss Institute of Bioinformatics, University of Zurich, Zurich, Switzerland
Christian von Mering
You can also search for this author in PubMed Google Scholar
PCAWG Evolution & Heterogeneity Working Group
- Stefan C. Dentro
- , Ignaty Leshchiner
- , Moritz Gerstung
- , Clemency Jolly
- , Kerstin Haase
- , Maxime Tarabichi
- , Jeff Wintersinger
- , Amit G. Deshwar
- , Kaixian Yu
- , Santiago Gonzalez
- , Yulia Rubanova
- , Geoff Macintyre
- , David J. Adams
- , Pavana Anur
- , Rameen Beroukhim
- , Paul C. Boutros
- , David D. Bowtell
- , Peter J. Campbell
- , Shaolong Cao
- , Elizabeth L. Christie
- , Marek Cmero
- , Yupeng Cun
- , Kevin J. Dawson
- , Jonas Demeulemeester
- , Nilgun Donmez
- , Ruben M. Drews
- , Roland Eils
- , Matthew Fittall
- , Dale W. Garsed
- , Marcin Imielinski
- , Lara Jerman
- , Kortine Kleinheinz
- , Juhee Lee
- , Henry Lee-Six
- , Dimitri G. Livitz
- , Salem Malikic
- , Florian Markowetz
- , Inigo Martincorena
- , Thomas J. Mitchell
- , Ville Mustonen
- , Layla Oesper
- , Martin Peifer
- , Myron Peto
- , Benjamin J. Raphael
- , Daniel Rosebrock
- , S. Cenk Sahinalp
- , Adriana Salcedo
- , Matthias Schlesner
- , Steven Schumacher
- , Subhajit Sengupta
- , Ruian Shi
- , Seung Jun Shin
- , Oliver Spiro
- , Lincoln D. Stein
- , Ignacio Vázquez-García
- , Shankar Vembu
- , David A. Wheeler
- , Tsun-Po Yang
- , Xiaotong Yao
- , Hongtu Zhu
- , Wenyi Wang
- , Quaid D. Morris
- , Paul T. Spellman
- , David C. Wedge
- & Peter Van Loo
- , Federico Abascal
- , Adam Abeshouse
- , Hiroyuki Aburatani
- , Nishant Agrawal
- , Keun Soo Ahn
- , Sung-Min Ahn
- , Hiroshi Aikata
- , Rehan Akbani
- , Kadir C. Akdemir
- , Hikmat Al-Ahmadie
- , Sultan T. Al-Sedairy
- , Fatima Al-Shahrour
- , Malik Alawi
- , Monique Albert
- , Kenneth Aldape
- , Ludmil B. Alexandrov
- , Adrian Ally
- , Kathryn Alsop
- , Eva G. Alvarez
- , Fernanda Amary
- , Samirkumar B. Amin
- , Brice Aminou
- , Ole Ammerpohl
- , Matthew J. Anderson
- , Davide Antonello
- , Samuel Aparicio
- , Elizabeth L. Appelbaum
- , Yasuhito Arai
- , Axel Aretz
- , Koji Arihiro
- , Shun-ichi Ariizumi
- , Joshua Armenia
- , Laurent Arnould
- , Sylvia Asa
- , Yassen Assenov
- , Gurnit Atwal
- , Sietse Aukema
- , J. Todd Auman
- , Miriam R. R. Aure
- , Philip Awadalla
- , Marta Aymerich
- , Gary D. Bader
- , Adrian Baez-Ortega
- , Matthew H. Bailey
- , Peter J. Bailey
- , Miruna Balasundaram
- , Saianand Balu
- , Pratiti Bandopadhayay
- , Rosamonde E. Banks
- , Stefano Barbi
- , Andrew P. Barbour
- , Jonathan Barenboim
- , Jill Barnholtz-Sloan
- , Hugh Barr
- , Elisabet Barrera
- , John Bartlett
- , Javier Bartolome
- , Claudio Bassi
- , Oliver F. Bathe
- , Daniel Baumhoer
- , Prashant Bavi
- , Stephen B. Baylin
- , Wojciech Bazant
- , Duncan Beardsmore
- , Timothy A. Beck
- , Sam Behjati
- , Andreas Behren
- , Beifang Niu
- , Cindy Bell
- , Sergi Beltran
- , Christopher Benz
- , Andrew Berchuck
- , Anke K. Bergmann
- , Erik N. Bergstrom
- , Benjamin P. Berman
- , Daniel M. Berney
- , Stephan H. Bernhart
- , Mario Berrios
- , Samantha Bersani
- , Johanna Bertl
- , Miguel Betancourt
- , Vinayak Bhandari
- , Shriram G. Bhosle
- , Andrew V. Biankin
- , Matthias Bieg
- , Darell Bigner
- , Hans Binder
- , Ewan Birney
- , Michael Birrer
- , Nidhan K. Biswas
- , Bodil Bjerkehagen
- , Tom Bodenheimer
- , Lori Boice
- , Giada Bonizzato
- , Johann S. De Bono
- , Arnoud Boot
- , Moiz S. Bootwalla
- , Arndt Borkhardt
- , Keith A. Boroevich
- , Ivan Borozan
- , Christoph Borst
- , Marcus Bosenberg
- , Mattia Bosio
- , Jacqueline Boultwood
- , Guillaume Bourque
- , G. Steven Bova
- , David T. Bowen
- , Reanne Bowlby
- , David D. L. Bowtell
- , Sandrine Boyault
- , Rich Boyce
- , Jeffrey Boyd
- , Alvis Brazma
- , Paul Brennan
- , Daniel S. Brewer
- , Arie B. Brinkman
- , Robert G. Bristow
- , Russell R. Broaddus
- , Jane E. Brock
- , Malcolm Brock
- , Annegien Broeks
- , Angela N. Brooks
- , Denise Brooks
- , Benedikt Brors
- , Søren Brunak
- , Timothy J. C. Bruxner
- , Alicia L. Bruzos
- , Alex Buchanan
- , Ivo Buchhalter
- , Christiane Buchholz
- , Susan Bullman
- , Hazel Burke
- , Birgit Burkhardt
- , Kathleen H. Burns
- , John Busanovich
- , Carlos D. Bustamante
- , Adam P. Butler
- , Atul J. Butte
- , Niall J. Byrne
- , Anne-Lise Børresen-Dale
- , Samantha J. Caesar-Johnson
- , Andy Cafferkey
- , Declan Cahill
- , Claudia Calabrese
- , Carlos Caldas
- , Fabien Calvo
- , Niedzica Camacho
- , Elias Campo
- , Cinzia Cantù
- , Thomas E. Carey
- , Joana Carlevaro-Fita
- , Rebecca Carlsen
- , Ivana Cataldo
- , Mario Cazzola
- , Jonathan Cebon
- , Robert Cerfolio
- , Dianne E. Chadwick
- , Dimple Chakravarty
- , Don Chalmers
- , Calvin Wing Yiu Chan
- , Michelle Chan-Seng-Yue
- , Vishal S. Chandan
- , David K. Chang
- , Stephen J. Chanock
- , Lorraine A. Chantrill
- , Aurélien Chateigner
- , Nilanjan Chatterjee
- , Kazuaki Chayama
- , Hsiao-Wei Chen
- , Jieming Chen
- , Yiwen Chen
- , Zhaohong Chen
- , Andrew D. Cherniack
- , Jeremy Chien
- , Yoke-Eng Chiew
- , Suet-Feung Chin
- , Sunghoon Cho
- , Jung Kyoon Choi
- , Christine Chomienne
- , Zechen Chong
- , Su Pin Choo
- , Angela Chou
- , Angelika N. Christ
- , Eric Chuah
- , Carrie Cibulskis
- , Kristian Cibulskis
- , Sara Cingarlini
- , Peter Clapham
- , Alexander Claviez
- , Sean Cleary
- , Nicole Cloonan
- , Colin C. Collins
- , Ashton A. Connor
- , Susanna L. Cooke
- , Colin S. Cooper
- , Leslie Cope
- , Vincenzo Corbo
- , Matthew G. Cordes
- , Stephen M. Cordner
- , Isidro Cortés-Ciriano
- , Kyle Covington
- , Prue A. Cowin
- , Brian Craft
- , David Craft
- , Chad J. Creighton
- , Erin Curley
- , Ioana Cutcutache
- , Karolina Czajka
- , Bogdan Czerniak
- , Rebecca A. Dagg
- , Ludmila Danilova
- , Maria Vittoria Davi
- , Natalie R. Davidson
- , Helen Davies
- , Ian J. Davis
- , Brandi N. Davis-Dusenbery
- , Francisco M. De La Vega
- , Ricardo De Paoli-Iseppi
- , Timothy Defreitas
- , Angelo P. Dei Tos
- , Olivier Delaneau
- , John A. Demchok
- , German M. Demidov
- , Deniz Demircioğlu
- , Nening M. Dennis
- , Robert E. Denroche
- , Stefan C. Dentro
- , Nikita Desai
- , Vikram Deshpande
- , Christine Desmedt
- , Jordi Deu-Pons
- , Noreen Dhalla
- , Neesha C. Dhani
- , Priyanka Dhingra
- , Rajiv Dhir
- , Anthony DiBiase
- , Klev Diamanti
- , Shuai Ding
- , Huy Q. Dinh
- , Luc Dirix
- , HarshaVardhan Doddapaneni
- , Michelle T. Dow
- , Ronny Drapkin
- , Oliver Drechsel
- , Serge Serge
- , Tim Dudderidge
- , Ana Dueso-Barroso
- , Andrew J. Dunford
- , Michael Dunn
- , Lewis Jonathan Dursi
- , Fraser R. Duthie
- , Ken Dutton-Regester
- , Jenna Eagles
- , Douglas F. Easton
- , Stuart Edmonds
- , Paul A. Edwards
- , Sandra E. Edwards
- , Rosalind A. Eeles
- , Anna Ehinger
- , Juergen Eils
- , Adel El-Naggar
- , Matthew Eldridge
- , Kyle Ellrott
- , Serap Erkek
- , Georgia Escaramis
- , Shadrielle M. G. Espiritu
- , Xavier Estivill
- , Dariush Etemadmoghadam
- , Jorunn E. Eyfjord
- , Bishoy M. Faltas
- , Daiming Fan
- , William C. Faquin
- , Claudiu Farcas
- , Matteo Fassan
- , Aquila Fatima
- , Francesco Favero
- , Nodirjon Fayzullaev
- , Ina Felau
- , Sian Fereday
- , Martin L. Ferguson
- , Vincent Ferretti
- , Lars Feuerbach
- , Matthew A. Field
- , J. Lynn Fink
- , Gaetano Finocchiaro
- , Cyril Fisher
- , Matthew W. Fittall
- , Anna Fitzgerald
- , Rebecca C. Fitzgerald
- , Adrienne M. Flanagan
- , Neil E. Fleshner
- , Paul Flicek
- , John A. Foekens
- , Kwun M. Fong
- , Nuno A. Fonseca
- , Christopher S. Foster
- , Natalie S. Fox
- , Michael Fraser
- , Scott Frazer
- , Milana Frenkel-Morgenstern
- , William Friedman
- , Joan Frigola
- , Catrina C. Fronick
- , Akihiro Fujimoto
- , Masashi Fujita
- , Masashi Fukayama
- , Lucinda A. Fulton
- , Robert S. Fulton
- , Mayuko Furuta
- , P. Andrew Futreal
- , Anja Füllgrabe
- , Stacey B. Gabriel
- , Steven Gallinger
- , Carlo Gambacorti-Passerini
- , Jianjiong Gao
- , Shengjie Gao
- , Levi Garraway
- , Øystein Garred
- , Erik Garrison
- , Nils Gehlenborg
- , Josep L. L. Gelpi
- , Joshy George
- , Daniela S. Gerhard
- , Clarissa Gerhauser
- , Jeffrey E. Gershenwald
- , Mark Gerstein
- , Mohammed Ghori
- , Ronald Ghossein
- , Nasra H. Giama
- , Richard A. Gibbs
- , Bob Gibson
- , Anthony J. Gill
- , Pelvender Gill
- , Dilip D. Giri
- , Dominik Glodzik
- , Vincent J. Gnanapragasam
- , Maria Elisabeth Goebler
- , Mary J. Goldman
- , Carmen Gomez
- , Abel Gonzalez-Perez
- , Dmitry A. Gordenin
- , James Gossage
- , Kunihito Gotoh
- , Ramaswamy Govindan
- , Dorthe Grabau
- , Janet S. Graham
- , Robert C. Grant
- , Anthony R. Green
- , Eric Green
- , Liliana Greger
- , Nicola Grehan
- , Sonia Grimaldi
- , Sean M. Grimmond
- , Robert L. Grossman
- , Adam Grundhoff
- , Gunes Gundem
- , Qianyun Guo
- , Manaswi Gupta
- , Shailja Gupta
- , Ivo G. Gut
- , Marta Gut
- , Jonathan Göke
- , Andrea Haake
- , David Haan
- , Siegfried Haas
- , James E. Haber
- , Nina Habermann
- , Faraz Hach
- , Syed Haider
- , Natsuko Hama
- , Freddie C. Hamdy
- , Anne Hamilton
- , Mark P. Hamilton
- , George B. Hanna
- , Martin Hansmann
- , Nicholas J. Haradhvala
- , Olivier Harismendy
- , Ivon Harliwong
- , Arif O. Harmanci
- , Eoghan Harrington
- , Takanori Hasegawa
- , David Haussler
- , Steve Hawkins
- , Shinya Hayami
- , Shuto Hayashi
- , D. Neil Hayes
- , Stephen J. Hayes
- , Nicholas K. Hayward
- , Steven Hazell
- , Allison P. Heath
- , Simon C. Heath
- , David Hedley
- , Apurva M. Hegde
- , David I. Heiman
- , Michael C. Heinold
- , Zachary Heins
- , Lawrence E. Heisler
- , Eva Hellstrom-Lindberg
- , Mohamed Helmy
- , Seong Gu Heo
- , Austin J. Hepperla
- , José María Heredia-Genestar
- , Carl Herrmann
- , Peter Hersey
- , Julian M. Hess
- , Holmfridur Hilmarsdottir
- , Jonathan Hinton
- , Satoshi Hirano
- , Nobuyoshi Hiraoka
- , Katherine A. Hoadley
- , Asger Hobolth
- , Ermin Hodzic
- , Jessica I. Hoell
- , Steve Hoffmann
- , Oliver Hofmann
- , Andrea Holbrook
- , Aliaksei Z. Holik
- , Michael A. Hollingsworth
- , Oliver Holmes
- , Robert A. Holt
- , Chen Hong
- , Eun Pyo Hong
- , Jongwhi H. Hong
- , Gerrit K. Hooijer
- , Henrik Hornshøj
- , Fumie Hosoda
- , Volker Hovestadt
- , William Howat
- , Alan P. Hoyle
- , Ralph H. Hruban
- , Jianhong Hu
- , Kuan-lin Huang
- , Mei Huang
- , Mi Ni Huang
- , Vincent Huang
- , Wolfgang Huber
- , Thomas J. Hudson
- , Michael Hummel
- , Jillian A. Hung
- , David Huntsman
- , Ted R. Hupp
- , Jason Huse
- , Matthew R. Huska
- , Barbara Hutter
- , Carolyn M. Hutter
- , Daniel Hübschmann
- , Christine A. Iacobuzio-Donahue
- , Charles David Imbusch
- , Seiya Imoto
- , William B. Isaacs
- , Keren Isaev
- , Shumpei Ishikawa
- , Murat Iskar
- , S. M. Ashiqul Islam
- , Michael Ittmann
- , Sinisa Ivkovic
- , Jose M. G. Izarzugaza
- , Jocelyne Jacquemier
- , Valerie Jakrot
- , Nigel B. Jamieson
- , Gun Ho Jang
- , Se Jin Jang
- , Joy C. Jayaseelan
- , Reyka Jayasinghe
- , Stuart R. Jefferys
- , Karine Jegalian
- , Jennifer L. Jennings
- , Seung-Hyup Jeon
- , Peter A. Johansson
- , Amber L. Johns
- , Jeremy Johns
- , Rory Johnson
- , Todd A. Johnson
- , Yann Joly
- , Jon G. Jonasson
- , Corbin D. Jones
- , David R. Jones
- , David T. W. Jones
- , Nic Jones
- , Steven J. M. Jones
- , Jos Jonkers
- , Young Seok Ju
- , Hartmut Juhl
- , Jongsun Jung
- , Malene Juul
- , Randi Istrup Juul
- , Sissel Juul
- , Natalie Jäger
- , Rolf Kabbe
- , Andre Kahles
- , Abdullah Kahraman
- , Vera B. Kaiser
- , Hojabr Kakavand
- , Sangeetha Kalimuthu
- , Christof von Kalle
- , Koo Jeong Kang
- , Katalin Karaszi
- , Beth Karlan
- , Rosa Karlić
- , Dennis Karsch
- , Katayoon Kasaian
- , Karin S. Kassahn
- , Hitoshi Katai
- , Mamoru Kato
- , Hiroto Katoh
- , Yoshiiku Kawakami
- , Jonathan D. Kay
- , Stephen H. Kazakoff
- , Marat D. Kazanov
- , Maria Keays
- , Electron Kebebew
- , Richard F. Kefford
- , Manolis Kellis
- , James G. Kench
- , Catherine J. Kennedy
- , Jules N. A. Kerssemakers
- , David Khoo
- , Vincent Khoo
- , Narong Khuntikeo
- , Ekta Khurana
- , Helena Kilpinen
- , Hark Kyun Kim
- , Hyung-Lae Kim
- , Hyung-Yong Kim
- , Hyunghwan Kim
- , Jaegil Kim
- , Jihoon Kim
- , Jong K. Kim
- , Youngwook Kim
- , Tari A. King
- , Wolfram Klapper
- , Leszek J. Klimczak
- , Stian Knappskog
- , Michael Kneba
- , Bartha M. Knoppers
- , Youngil Koh
- , Jan Komorowski
- , Daisuke Komura
- , Mitsuhiro Komura
- , Marcel Kool
- , Jan O. Korbel
- , Viktoriya Korchina
- , Andrey Korshunov
- , Michael Koscher
- , Roelof Koster
- , Zsofia Kote-Jarai
- , Antonios Koures
- , Milena Kovacevic
- , Barbara Kremeyer
- , Helene Kretzmer
- , Markus Kreuz
- , Savitri Krishnamurthy
- , Dieter Kube
- , Kiran Kumar
- , Pardeep Kumar
- , Sushant Kumar
- , Yogesh Kumar
- , Ritika Kundra
- , Kirsten Kübler
- , Ralf Küppers
- , Jesper Lagergren
- , Phillip H. Lai
- , Peter W. Laird
- , Sunil R. Lakhani
- , Christopher M. Lalansingh
- , Emilie Lalonde
- , Fabien C. Lamaze
- , Adam Lambert
- , Eric Lander
- , Pablo Landgraf
- , Luca Landoni
- , Anita Langerød
- , Andrés Lanzós
- , Denis Larsimont
- , Erik Larsson
- , Mark Lathrop
- , Loretta M. S. Lau
- , Chris Lawerenz
- , Rita T. Lawlor
- , Michael S. Lawrence
- , Alexander J. Lazar
- , Ana Mijalkovic Lazic
- , Darlene Lee
- , Donghoon Lee
- , Eunjung Alice Lee
- , Hee Jin Lee
- , Jake June-Koo Lee
- , Jeong-Yeon Lee
- , Ming Ta Michael Lee
- , Kjong-Van Lehmann
- , Hans Lehrach
- , Dido Lenze
- , Conrad R. Leonard
- , Daniel A. Leongamornlert
- , Louis Letourneau
- , Ivica Letunic
- , Douglas A. Levine
- , Lora Lewis
- , Constance H. Li
- , Haiyan Irene Li
- , Shantao Li
- , Siliang Li
- , Xiaobo Li
- , Xiaotong Li
- , Xinyue Li
- , Yilong Li
- , Han Liang
- , Sheng-Ben Liang
- , Peter Lichter
- , W. M. Linehan
- , Ole Christian Lingjærde
- , Dongbing Liu
- , Eric Minwei Liu
- , Fei-Fei Fei Liu
- , Fenglin Liu
- , Xingmin Liu
- , Julie Livingstone
- , Dimitri Livitz
- , Naomi Livni
- , Lucas Lochovsky
- , Markus Loeffler
- , Georgina V. Long
- , Armando Lopez-Guillermo
- , Shaoke Lou
- , David N. Louis
- , Laurence B. Lovat
- , Yiling Lu
- , Yong-Jie Lu
- , Youyong Lu
- , Claudio Luchini
- , Ilinca Lungu
- , Xuemei Luo
- , Hayley J. Luxton
- , Andy G. Lynch
- , Lisa Lype
- , Cristina López
- , Carlos López-Otín
- , Eric Z. Ma
- , Yussanne Ma
- , Gaetan MacGrogan
- , Shona MacRae
- , Tobias Madsen
- , Kazuhiro Maejima
- , Andrea Mafficini
- , Dennis T. Maglinte
- , Arindam Maitra
- , Partha P. Majumder
- , Luca Malcovati
- , Giuseppe Malleo
- , Graham J. Mann
- , Luisa Mantovani-Löffler
- , Kathleen Marchal
- , Giovanni Marchegiani
- , Elaine R. Mardis
- , Adam A. Margolin
- , Maximillian G. Marin
- , Julia Markowski
- , Jeffrey Marks
- , Tomas Marques-Bonet
- , Marco A. Marra
- , Luke Marsden
- , John W. M. Martens
- , Sancha Martin
- , Jose I. Martin-Subero
- , Iñigo Martincorena
- , Alexander Martinez-Fundichely
- , Yosef E. Maruvka
- , R. Jay Mashl
- , Charlie E. Massie
- , Thomas J. Matthew
- , Lucy Matthews
- , Erik Mayer
- , Simon Mayes
- , Michael Mayo
- , Faridah Mbabaali
- , Karen McCune
- , Ultan McDermott
- , Patrick D. McGillivray
- , Michael D. McLellan
- , John D. McPherson
- , John R. McPherson
- , Treasa A. McPherson
- , Samuel R. Meier
- , Alice Meng
- , Shaowu Meng
- , Andrew Menzies
- , Neil D. Merrett
- , Sue Merson
- , Matthew Meyerson
- , William Meyerson
- , Piotr A. Mieczkowski
- , George L. Mihaiescu
- , Sanja Mijalkovic
- , Tom Mikkelsen
- , Michele Milella
- , Linda Mileshkin
- , Christopher A. Miller
- , David K. Miller
- , Jessica K. Miller
- , Gordon B. Mills
- , Ana Milovanovic
- , Sarah Minner
- , Marco Miotto
- , Gisela Mir Arnau
- , Lisa Mirabello
- , Chris Mitchell
- , Satoru Miyano
- , Naoki Miyoshi
- , Shinichi Mizuno
- , Fruzsina Molnár-Gábor
- , Malcolm J. Moore
- , Richard A. Moore
- , Sandro Morganella
- , Carl Morrison
- , Lisle E. Mose
- , Catherine D. Moser
- , Ferran Muiños
- , Loris Mularoni
- , Andrew J. Mungall
- , Karen Mungall
- , Elizabeth A. Musgrove
- , David Mutch
- , Francesc Muyas
- , Donna M. Muzny
- , Alfonso Muñoz
- , Jerome Myers
- , Ola Myklebost
- , Peter Möller
- , Genta Nagae
- , Adnan M. Nagrial
- , Hardeep K. Nahal-Bose
- , Hitoshi Nakagama
- , Hidewaki Nakagawa
- , Hiromi Nakamura
- , Toru Nakamura
- , Kaoru Nakano
- , Tannistha Nandi
- , Jyoti Nangalia
- , Mia Nastic
- , Arcadi Navarro
- , Fabio C. P. Navarro
- , David E. Neal
- , Gerd Nettekoven
- , Felicity Newell
- , Steven J. Newhouse
- , Yulia Newton
- , Alvin Wei Tian Ng
- , Anthony Ng
- , Jonathan Nicholson
- , David Nicol
- , Yongzhan Nie
- , G. Petur Nielsen
- , Morten Muhlig Nielsen
- , Serena Nik-Zainal
- , Michael S. Noble
- , Katia Nones
- , Paul A. Northcott
- , Faiyaz Notta
- , Brian D. O’Connor
- , Peter O’Donnell
- , Maria O’Donovan
- , Sarah O’Meara
- , Brian Patrick O’Neill
- , J. Robert O’Neill
- , David Ocana
- , Angelica Ochoa
- , Christopher Ogden
- , Hideki Ohdan
- , Kazuhiro Ohi
- , Lucila Ohno-Machado
- , Karin A. Oien
- , Akinyemi I. Ojesina
- , Hidenori Ojima
- , Takuji Okusaka
- , Larsson Omberg
- , Choon Kiat Ong
- , Stephan Ossowski
- , German Ott
- , B. F. Francis Ouellette
- , Christine P’ng
- , Marta Paczkowska
- , Salvatore Paiella
- , Chawalit Pairojkul
- , Marina Pajic
- , Qiang Pan-Hammarström
- , Elli Papaemmanuil
- , Irene Papatheodorou
- , Nagarajan Paramasivam
- , Ji Wan Park
- , Joong-Won Park
- , Keunchil Park
- , Kiejung Park
- , Peter J. Park
- , Joel S. Parker
- , Simon L. Parsons
- , Harvey Pass
- , Danielle Pasternack
- , Alessandro Pastore
- , Ann-Marie Patch
- , Iris Pauporté
- , Antonio Pea
- , John V. Pearson
- , Chandra Sekhar Pedamallu
- , Jakob Skou Pedersen
- , Paolo Pederzoli
- , Nathan A. Pennell
- , Charles M. Perou
- , Marc D. Perry
- , Gloria M. Petersen
- , Nicholas Petrelli
- , Robert Petryszak
- , Stefan M. Pfister
- , Mark Phillips
- , Oriol Pich
- , Hilda A. Pickett
- , Todd D. Pihl
- , Nischalan Pillay
- , Sarah Pinder
- , Mark Pinese
- , Andreia V. Pinho
- , Esa Pitkänen
- , Xavier Pivot
- , Elena Piñeiro-Yáñez
- , Laura Planko
- , Christoph Plass
- , Paz Polak
- , Tirso Pons
- , Irinel Popescu
- , Olga Potapova
- , Aparna Prasad
- , Shaun R. Preston
- , Manuel Prinz
- , Antonia L. Pritchard
- , Stephenie D. Prokopec
- , Elena Provenzano
- , Xose S. Puente
- , Sonia Puig
- , Montserrat Puiggròs
- , Sergio Pulido-Tamayo
- , Gulietta M. Pupo
- , Colin A. Purdie
- , Michael C. Quinn
- , Raquel Rabionet
- , Janet S. Rader
- , Bernhard Radlwimmer
- , Petar Radovic
- , Benjamin Raeder
- , Keiran M. Raine
- , Manasa Ramakrishna
- , Kamna Ramakrishnan
- , Suresh Ramalingam
- , W. Kimryn Rathmell
- , Tobias Rausch
- , Guido Reifenberger
- , Jüri Reimand
- , Jorge Reis-Filho
- , Victor Reuter
- , Iker Reyes-Salazar
- , Matthew A. Reyna
- , Sheila M. Reynolds
- , Esther Rheinbay
- , Yasser Riazalhosseini
- , Andrea L. Richardson
- , Julia Richter
- , Matthew Ringel
- , Markus Ringnér
- , Yasushi Rino
- , Karsten Rippe
- , Jeffrey Roach
- , Lewis R. Roberts
- , Nicola D. Roberts
- , Steven A. Roberts
- , A. Gordon Robertson
- , Alan J. Robertson
- , Javier Bartolomé Rodriguez
- , Bernardo Rodriguez-Martin
- , F. Germán Rodríguez-González
- , Michael H. A. Roehrl
- , Marius Rohde
- , Hirofumi Rokutan
- , Gilles Romieu
- , Ilse Rooman
- , Tom Roques
- , Mara Rosenberg
- , Philip C. Rosenstiel
- , Andreas Rosenwald
- , Edward W. Rowe
- , Romina Royo
- , Steven G. Rozen
- , Mark A. Rubin
- , Carlota Rubio-Perez
- , Vasilisa A. Rudneva
- , Borislav C. Rusev
- , Andrea Ruzzenente
- , Gunnar Rätsch
- , Radhakrishnan Sabarinathan
- , Veronica Y. Sabelnykova
- , Sara Sadeghi
- , Natalie Saini
- , Mihoko Saito-Adachi
- , Gordon Saksena
- , Roberto Salgado
- , Leonidas Salichos
- , Richard Sallari
- , Charles Saller
- , Roberto Salvia
- , Michelle Sam
- , Jaswinder S. Samra
- , Francisco Sanchez-Vega
- , Chris Sander
- , Grant Sanders
- , Rajiv Sarin
- , Iman Sarrafi
- , Aya Sasaki-Oku
- , Torill Sauer
- , Guido Sauter
- , Robyn P. M. Saw
- , Maria Scardoni
- , Christopher J. Scarlett
- , Aldo Scarpa
- , Ghislaine Scelo
- , Dirk Schadendorf
- , Jacqueline E. Schein
- , Markus B. Schilhabel
- , Thorsten Schlomm
- , Heather K. Schmidt
- , Sarah-Jane Schramm
- , Stefan Schreiber
- , Nikolaus Schultz
- , Steven E. Schumacher
- , Roland F. Schwarz
- , Richard A. Scolyer
- , David Scott
- , Ralph Scully
- , Raja Seethala
- , Ayellet V. Segre
- , Iris Selander
- , Colin A. Semple
- , Yasin Senbabaoglu
- , Elisabetta Sereni
- , Stefano Serra
- , Dennis C. Sgroi
- , Mark Shackleton
- , Nimish C. Shah
- , Sagedeh Shahabi
- , Catherine A. Shang
- , Ping Shang
- , Ofer Shapira
- , Troy Shelton
- , Ciyue Shen
- , Rebecca Shepherd
- , Yu-Jia Shiah
- , Tatsuhiro Shibata
- , Juliann Shih
- , Eigo Shimizu
- , Kiyo Shimizu
- , Yuichi Shiraishi
- , Tal Shmaya
- , Ilya Shmulevich
- , Solomon I. Shorser
- , Charles Short
- , Raunak Shrestha
- , Suyash S. Shringarpure
- , Craig Shriver
- , Shimin Shuai
- , Nikos Sidiropoulos
- , Reiner Siebert
- , Anieta M. Sieuwerts
- , Lina Sieverling
- , Sabina Signoretti
- , Katarzyna O. Sikora
- , Michele Simbolo
- , Ronald Simon
- , Janae V. Simons
- , Jared T. Simpson
- , Peter T. Simpson
- , Samuel Singer
- , Nasa Sinnott-Armstrong
- , Payal Sipahimalani
- , Tara J. Skelly
- , Marcel Smid
- , Jaclyn Smith
- , Karen Smith-McCune
- , Nicholas D. Socci
- , Heidi J. Sofia
- , Matthew G. Soloway
- , Anil K. Sood
- , Sharmila Sothi
- , Christos Sotiriou
- , Cameron M. Soulette
- , Paul N. Span
- , Nicola Sperandio
- , Andrew J. Spillane
- , Jonathan Spring
- , Johan Staaf
- , Peter F. Stadler
- , Peter Staib
- , Stefan G. Stark
- , Lucy Stebbings
- , Ólafur Andri Stefánsson
- , Oliver Stegle
- , Alasdair Stenhouse
- , Chip Stewart
- , Stephan Stilgenbauer
- , Miranda D. Stobbe
- , Michael R. Stratton
- , Jonathan R. Stretch
- , Adam J. Struck
- , Joshua M. Stuart
- , Henk G. Stunnenberg
- , Xiaoping Su
- , Ren X. Sun
- , Stephanie Sungalee
- , Hana Susak
- , Akihiro Suzuki
- , Fred Sweep
- , Monika Szczepanowski
- , Holger Sültmann
- , Takashi Yugawa
- , Angela Tam
- , David Tamborero
- , Benita Kiat Tee Tan
- , Donghui Tan
- , Patrick Tan
- , Hiroko Tanaka
- , Hirokazu Taniguchi
- , Tomas J. Tanskanen
- , Roy Tarnuzzer
- , Patrick Tarpey
- , Morgan L. Taschuk
- , Kenji Tatsuno
- , Simon Tavaré
- , Darrin F. Taylor
- , Amaro Taylor-Weiner
- , Jon W. Teague
- , Bin Tean Teh
- , Varsha Tembe
- , Javier Temes
- , Kevin Thai
- , Sarah P. Thayer
- , Nina Thiessen
- , Gilles Thomas
- , Sarah Thomas
- , Alan Thompson
- , Alastair M. Thompson
- , John F. F. Thompson
- , R. Houston Thompson
- , Heather Thorne
- , Leigh B. Thorne
- , Adrian Thorogood
- , Grace Tiao
- , Nebojsa Tijanic
- , Lee E. Timms
- , Roberto Tirabosco
- , Marta Tojo
- , Stefania Tommasi
- , Christopher W. Toon
- , Umut H. Toprak
- , David Torrents
- , Giampaolo Tortora
- , Jörg Tost
- , Yasushi Totoki
- , David Townend
- , Nadia Traficante
- , Isabelle Treilleux
- , Jean-Rémi Trotta
- , Lorenz H. P. Trümper
- , Ming Tsao
- , Tatsuhiko Tsunoda
- , Jose M. C. Tubio
- , Olga Tucker
- , Richard Turkington
- , Daniel J. Turner
- , Andrew Tutt
- , Masaki Ueno
- , Naoto T. Ueno
- , Christopher Umbricht
- , Husen M. Umer
- , Timothy J. Underwood
- , Lara Urban
- , Tomoko Urushidate
- , Tetsuo Ushiku
- , Liis Uusküla-Reimand
- , Alfonso Valencia
- , David J. Van Den Berg
- , Steven Van Laere
- , Peter Van Loo
- , Erwin G. Van Meir
- , Gert G. Van den Eynden
- , Theodorus Van der Kwast
- , Naveen Vasudev
- , Miguel Vazquez
- , Ravikiran Vedururu
- , Umadevi Veluvolu
- , Lieven P. C. Verbeke
- , Peter Vermeulen
- , Clare Verrill
- , Alain Viari
- , David Vicente
- , Caterina Vicentini
- , K. VijayRaghavan
- , Juris Viksna
- , Ricardo E. Vilain
- , Izar Villasante
- , Anne Vincent-Salomon
- , Tapio Visakorpi
- , Douglas Voet
- , Paresh Vyas
- , Nick M. Waddell
- , Nicola Waddell
- , Claes Wadelius
- , Lina Wadi
- , Rabea Wagener
- , Jeremiah A. Wala
- , Jian Wang
- , Jiayin Wang
- , Linghua Wang
- , Yumeng Wang
- , Zhining Wang
- , Paul M. Waring
- , Hans-Jörg Warnatz
- , Jonathan Warrell
- , Anne Y. Warren
- , Sebastian M. Waszak
- , Dieter Weichenhan
- , Paul Weinberger
- , John N. Weinstein
- , Joachim Weischenfeldt
- , Daniel J. Weisenberger
- , Ian Welch
- , Michael C. Wendl
- , Johannes Werner
- , Justin P. Whalley
- , Hayley C. Whitaker
- , Dennis Wigle
- , Matthew D. Wilkerson
- , Ashley Williams
- , James S. Wilmott
- , Gavin W. Wilson
- , Julie M. Wilson
- , Richard K. Wilson
- , Boris Winterhoff
- , Jeffrey A. Wintersinger
- , Maciej Wiznerowicz
- , Stephan Wolf
- , Bernice H. Wong
- , Tina Wong
- , Winghing Wong
- , Youngchoon Woo
- , Scott Wood
- , Bradly G. Wouters
- , Adam J. Wright
- , Derek W. Wright
- , Mark H. Wright
- , Chin-Lee Wu
- , Dai-Ying Wu
- , Guanming Wu
- , Jianmin Wu
- , Zhenggang Wu
- , Qian Xiang
- , Xiao Xiao
- , Heng Xiong
- , Qinying Xu
- , Yanxun Xu
- , Shinichi Yachida
- , Sergei Yakneen
- , Rui Yamaguchi
- , Takafumi N. Yamaguchi
- , Masakazu Yamamoto
- , Shogo Yamamoto
- , Hiroki Yamaue
- , Huanming Yang
- , Jean Y. Yang
- , Liming Yang
- , Lixing Yang
- , Shanlin Yang
- , Yang Yang
- , Marie-Laure Yaspo
- , Lucy Yates
- , Christina Yau
- , Venkata D. Yellapantula
- , Christopher J. Yoon
- , Sung-Soo Yoon
- , Fouad Yousif
- , Willie Yu
- , Yingyan Yu
- , Yuan Yuan
- , Denis Yuen
- , Christina K. Yung
- , Olga Zaikova
- , Jorge Zamora
- , Marc Zapatka
- , Jean C. Zenklusen
- , Thorsten Zenz
- , Nikolajs Zeps
- , Cheng-Zhong Zhang
- , Fan Zhang
- , Hailei Zhang
- , Hongwei Zhang
- , Hongxin Zhang
- , Jiashan Zhang
- , Jing Zhang
- , Junjun Zhang
- , Xiuqing Zhang
- , Xuanping Zhang
- , Yan Zhang
- , Zemin Zhang
- , Zhongming Zhao
- , Liangtao Zheng
- , Xiuqing Zheng
- , Wanding Zhou
- , Yong Zhou
- , Jingchun Zhu
- , Shida Zhu
- , Lihua Zou
- , Xueqing Zou
- , Anna deFazio
- , Nicholas van As
- , Carolien H. M. van Deurzen
- , Marc J. van de Vijver
- , L. van’t Veer
- & Christian von Mering
Contributions
M.G., C.J., I.L., S.G., P.A., D.R., D.G.L., P.T.S. and P.V.L. performed timing of point mutations and copy number gains. S.G. and M.G. performed qualitative timing of driver point mutations and analyses of synchronous gains, L.J. timed secondary copy number gains. I.L., T.J.M., D.R., D.G.L., D.C.W. and G.G. performed relative timing of somatic driver events and implemented integrative models. C.J., Y.R., M.G., Q.D.M. and P.V.L. performed timing of mutational signatures. M.G. performed real-time estimation of whole-genome duplication and subclonal diversification. S.G. assessed mutation rates in relapsed samples. C.J., M.G., I.L., Y.R., D.R. and P.V.L. constructed cancer timelines. M.G., C.J., I.L., S.C.D., S.G., T.J.M., Y.R., P.A., J.D., P.C.B., D.D.B., V.M., Q.D.M., P.T.S., D.C.W. and P.V.L. interpreted the results. S.C.D., I.L., J.W., A.D., I.V.-G., K. Yuan, G.M., M.P., S.M., N.D., K. Yu, S. Sengupta, K.H., M.T., J.D., D.G.L., D.R., J.L., M.C., S.C.S., Y.J., F.M., V.M., H.Z., W.W., Q.D.M., D.C.W. and P.V.L. performed subclonal architecture analysis. S.C.D., I.L., K.K., V.M., M.P., X.Y., D.G.L., S. Schumacher, R.B., M.I., M.S., D.C.W. and P.V.L. performed copy number analysis. J.W., S.C.D., I.L., K.H., D.G.L., K.K., D.R., D.C.W., Q.D.M. and P.V.L. derived a consensus of copy number analysis results. K. Yu, M.T., A.D., S.C.D., I.L., D.C.W., M.G., P.V.L., Q.D.M. and W.W. derived a consensus of subclonal architecture results. Y.F. and W.W. contributed to subclonal mutation calls. P.T.S., D.C.W. and P.V.L. coordinated the study. M.G., C.J., P.T.S., Y.R., I.L., Q.D.M., D.C.W. and P.V.L. wrote the manuscript, which all authors approved. S.C.D., I.L., M.G., C.J., K.H., M.T., J.W., A.G.D., K. Yu, S.G., Y.R. and G.M. in the PCAWG Evolution & Heterogeneity Working Group contributed equally. W.W., Q.D.M., P.T.S., D.C.W. and P.V.L. in the PCAWG Evolution & Heterogeneity Working Group jointly supervised the work.
Corresponding authors
Correspondence to Moritz Gerstung or Peter Van Loo .
Ethics declarations
Competing interests.
R.B. owns equity in Ampressa Therapeutics. G.G. receives research funds from IBM and Pharmacyclics and is an inventor on patent applications related to MuTect, ABSOLUTE, MutSig, MSMuTect and POLYSOLVER. I.L. is a consultant for PACT Pharma. B.J.R. is a consultant at and has ownership interest (including stock and patents) in Medley Genomics. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended data fig. 1 summary of all results obtained for colorectal adenocarcinoma ( n = 60) as an example..
a , Clustered heat maps of mutational timing estimates for gained segments, per patient. Colours as indicated in main text: green represents early clonal events, purple represents late clonal. b , Relative ordering of copy number events and driver mutations across all samples. c , Distribution of mutations across early clonal, late clonal and subclonal stages, for the most common driver genes. A maximum of 10 driver genes are shown. d , Clustered mutational signature fold changes between early clonal and late clonal stages, per patient. Green and purple indicate, respectively, a signature decrease and increase in late clonal from early clonal mutations. Inactive signatures are coloured white. e , As in d but for clonal versus subclonal stages. Blue indicates a signature decrease and red an increase in subclonal from clonal mutations. f , Typical timeline of tumour development. Similar result summaries for all other cancer types can be found in the Supplementary Information (pages 46–77).
Extended Data Fig. 2 Comparison of methods used for timing of individual copy number gains.
a , b , Pairwise comparison of the three approaches for timing individual copy number gains. c , Comparison using simulated data, showing high concordance.
Extended Data Fig. 3 Early copy number gains in brain cancers.
a , Three illustrative examples of glioblastoma with trisomy 7. The red arrow depicts the expected VAF cluster of point mutations preceding trisomy 7, which usually contains less than three SNVs. b , Distributions of the number of SNVs preceding trisomy 7 and total number of mutations on chromosome (chr) 7 in n = 34 GBM samples with trisomy 7. c , Medulloblastoma example with isochromosome 17q. d , Distributions of SNVs on 17q in n = 95 samples with isochromosome 17q; 74 out of 95 samples have less than 1 SNV preceding the isochromosome.
Extended Data Fig. 4 Validation of relative ordering model reconstruction based on simulated cohorts of whole-genome samples.
a , Relative ordering model (PhylogicNDT LeagueModel) results for a simulated cohort of samples ( n = 100) from a single generalized relative order of events (with varied prevalence) showing high concordance with the true trajectory. Probability distributions show the uncertainty of timing for specific events in the cohort. b , Relative ordering model results on a simulated cohort of samples ( n = 95) from a complex mixture of trajectories with different order of events showing high concordance with the expected average trajectory. c , Estimation of accuracy of the relative ordering model reconstruction by simulation of a set of 100 cohorts ( n (samples) = 100) with random trajectory mixtures and quantifying the distance in log odds early/late from perfect ordering. For the vast majority of events (even with low number of occurrences in the cohort), the log odds error does not exceed 1, confirming that very few events would switch between timing categories. The inset box corresponds to the first and third quartiles of the distribution, the horizontal line indicates the median and whiskers include data within 1.5× the IQR from the box. d , Simulated data show concordant timing in cohorts with WGD ( n = 245). Exclusion of samples with WGD (right, n = 242) introduces only a mild drop in accuracy, indicating that WGD is beneficial but not necessary for the reconstruction. Red dot = true rank. e , Estimated log odds in observed data including WGD (left, n = 245) and without (right, n = 242), across different mutation types. The inset box corresponds to the first and third quartiles of the distribution, the horizontal line indicates the median and whiskers include data within 1.5× the IQR from the box.
Extended Data Fig. 5 Correlation between the league model and Bradley–Terry model ordering.
Direct comparison for each tumour type of the league and Bradley–Terry models for determining the order of recurrent somatic mutations and copy number events. Axes indicate the ordered events observed in the respective tumour types. Correlation is quantified by Spearman’s rank correlation coefficient. A total of n = 756 ordered events are shown.
Extended Data Fig. 6 Examples of mutation spectrum changes across tumour evolution.
a , Three examples of tumours with substantial changes between mutation spectra of early (top) and late (bottom) clonal time points. b , Three examples of tumours with substantial changes between mutation spectra of clonal (top) and subclonal (bottom) time points.

Extended Data Fig. 7 Overview of early-to-late clonal and clonal-to-subclonal signature changes across tumour types.
a , b , Pie charts representing signature changes per cancer type for early-to-late clonal signature changes ( a ) and clonal-to-subclonal signature changes ( b ). Signatures that decrease between early and late are coloured green; signatures that increase are purple. The size of each pie chart represents the frequency of each signature. Signatures are split into three categories: (1) clock-like, comprising the putative clock signatures 1 and 5; (2) frequent, which are signatures present in ten or more cancer types; and (3) cancer-type specific, which are in fewer than ten cancer types and are often limited to specific cohorts.
Extended Data Fig. 8 Age-dependent mutation burden and relapse samples indicate near-normal CpG>TpG mutation rate in cancer, with moderate acceleration during carcinogenesis.
a , Across all cancer samples, a predominantly linear accumulation of CpG>TpG mutations (scaled to copy number) is observed over time, as measured by the age at diagnosis. b , Cancer-specific analysis of the CpG>TpG mutation burden as a function of age at diagnosis for n = 1,978 samples of 34 informative cancer types. The dotted line denotes the median mutations per year (that is, not offset), and shading denotes the 95% credible interval of a hierarchical Bayesian linear regression model across all data points. Slope and intercepts are drawn for each cancer type from a gamma distribution, respectively; inference was done by Hamiltonian Monte Carlo sampling. c , Maximum a posteriori estimates of rate and offset for 34 cancer types with 95% credible intervals as defined in b . d , Mutation rate inferred from cancer as in b and from selected normal tissue sequencing studies of n = 140 normal haematopoietic stem cells, n = 1 normal skin sample, n = 182 samples from normal endometrium, and n = 445 normal colonic crypts; error bars denote the 95% confidence interval. e , Median fraction of mutations attributed to linear age-dependent accumulation, based on estimates from b and the age at diagnosis for each sample. Error bars denote the 95% credible interval. f , g , CpG>TpG mutations per gigabase for ovarian cancer ( f ) and breast cancer ( g ) samples with matched primary and relapse samples. h , Increase in CpG>TpG mutation rate inferred from paired primary and relapse samples for six cancer types. Bars denote the range of the rate increase for different scenarios of copy number evolution, assuming ploidy changes have occurred prior (upper value) or posterior (lower value) to the branching between primary and relapse sample.
Extended Data Fig. 9 Real-time estimates indicate long latencies for some samples caused by the absence of early mutations.
a , Time of WGD for n = 571 individual patients, split by tumour type with an estimated mutation rate increase of 5×, except for ovary–adenocarcinoma (7.5×) and CNS (2.5×). Error bars represent 80% confidence intervals, reflecting uncertainty stemming from the number of mutations per segment and onset of the rate increase. Box plots demarcate the quartiles and median of the distribution with whiskers indicating 5% and 95% quantiles. b , Scatter plots showing the time of diagnosis ( x axis) and inferred time of WGD ( y axis) with error bars as in a . c , Scatter plot of early (co-amplified) CpG>TpG mutations ( y axis) as a function of the mutational time estimate of WGD ( x axis). The black line denotes a nonlinear loess fit with 95% confidence interval. Colours define the cancer type as in a . d , Total CpG>TpG mutations ( y axis) as a function of the mutation time estimate of WGD ( x axis). Colours and fit as in c . Early molecular timing is thus caused by a depletion of early CpG>TpG mutations, rather than an inflation of late CpG>TpG mutations. e , Estimated median WGD latency of n = 571 patients as in a for fixed ( x axis) versus patient specific rate increases, depending on the observed CpG>TpG mutation burden, allowing for a higher (up to 10×) mutation rate increase in samples with more mutations ( y axis). Error bars denote the IQR. f , Timing of subclonal diversification using CpG>TpG mutations in n = 1,953 individual patients. Box plots and error bars for data points as in a . g , Comparison of the median duration of subclonal diversification per cancer type assuming branching and linear phylogenies.
Supplementary information
Supplementary information.
This file contains a more detailed description of all methods, three supplementary notes, and summary pages for each PCAWG cohort, with sample-level figures representing the results of each of the life history analyses: timing of gains, ordering of events, timing of drivers, signature changes and evolutionary timelines.
Reporting Summary
PCAWG Consortium author list: This file contains a full list of consortium members.
Source Data Fig. 1
Source data fig. 2, source data fig. 3, source data fig. 4, source data fig. 5, source data fig. 6, source data extended data fig. 3, source data extended data fig. 5, source data extended data fig. 6, source data extended data fig. 8, source data extended data fig. 9, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Gerstung, M., Jolly, C., Leshchiner, I. et al. The evolutionary history of 2,658 cancers. Nature 578 , 122–128 (2020). https://doi.org/10.1038/s41586-019-1907-7
Download citation
Received : 11 August 2017
Accepted : 18 November 2019
Published : 05 February 2020
Issue Date : 06 February 2020
DOI : https://doi.org/10.1038/s41586-019-1907-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Computational validation of clonal and subclonal copy number alterations from bulk tumor sequencing using cnaqc.
- Alice Antonello
- Riccardo Bergamin
- Giulio Caravagna
Genome Biology (2024)
Evolving copy number gains promote tumor expansion and bolster mutational diversification
- Zicheng Wang
Nature Communications (2024)
Deep whole-genome analysis of 494 hepatocellular carcinomas
- Chong Zhang
- Hongyang Wang
Nature (2024)
ERK pathway agonism for cancer therapy: evidence, insights, and a target discovery framework
- Oleg Timofeev
- Philippe Giron
- Maxim Noeparast
npj Precision Oncology (2024)
CONIPHER: a computational framework for scalable phylogenetic reconstruction with error correction
- Kristiana Grigoriadis
- Ariana Huebner
- Nicholas McGranahan
Nature Protocols (2024)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Cancer newsletter — what matters in cancer research, free to your inbox weekly.
For the best browsing experience please enable JavaScript. Instructions for Microsoft Edge and Internet Explorer , other browsers

- About cancer
- Get involved
- Our research
- Funding for researchers
- Cancer types
- Cancer in general
- Causes of cancer
- Coping with cancer
- Health professionals
- Do your own fundraising
- By cancer type
- By cancer subject
- Our funding schemes
- Applying for funding
- Managing your research grant
- How we deliver our research
- Find a shop
- Shop online
- Our eBay shop
- Our organisation
- Current jobs
- Cancer news

A history of these pages
Cancer Research UK's About Cancer pages began life as CancerHelp UK. This was one of the first comprehensive cancer websites based in the UK.
Our pages include information about:
- the causes of cancer, how to reduce your risk and healthy living
- cancer symptoms and cancer screening
- cancer tests and treatments
- coping with a diagnosis of cancer and the treatment side effects
- research and clinical trials
The information is accessible, comprehensive and easy to read.
Co founders
The co founders of CancerHelp UK were Nick James and Sally Tweddle.
Nick James is a cancer specialist at the University Hospitals Birmingham NHS Foundation Trust. He is also a Professor of Oncology at the University of Birmingham Institute for Cancer Studies.
Sally Tweddle was an educationalist with an interest in literacy. Her husband's cancer and their search for cancer information online inspired CancerHelp UK.
To begin with, the Medical School server at the University of Birmingham hosted CancerHelp UK.
The management of the site passed to The Cancer Research Campaign in 2000. In 2002, The Cancer Research Campaign and the Imperial Cancer Research Fund merged. Together they formed Cancer Research UK. CancerHelp UK is now the About cancer section of Cancer Research UK's website.
Sarah Jane (Sally) Tweddle (1955–1999)
Sally was instrumental in setting up CancerHelp UK in 1994. At the time, most online cancer information was aimed at professionals. There was little information available for patients and relatives.
Sally’s background was in education. She saw how to present effective, detailed online material to the general public. The website launch was a success.
Sally earned a prestigious Fellowship in Cancer Education from the Cancer Research Campaign. In this role, she promoted the transmission of cancer information to the general public and to medical students.
Sadly, in 1999 Sally was diagnosed with a widespread cancer of the duodenum. She died peacefully at her home with her family around her, as she had wished.
Related links
About cancer research uk's patient and health information.
The history of Cancer Research UK's patient and health information website, how the information is produced and the processes we go through to make sure our information is accurate, up-to-date and engaging.
About cancer homepage
Find out about possible causes of cancer, how cancer starts and grows, tests to diagnose it, and general information about treatments.
It’s a worrying time for many people and we want to be there for you whenever - and wherever - you need us. Cancer Chat is our fully moderated forum where you can talk to others affected by cancer, share experiences, and get support. Cancer Chat is free to join and available 24 hours a day.
Visit the Cancer Chat forum
About Cancer generously supported by Dangoor Education since 2010.

Find a clinical trial
Search our clinical trials database for all cancer trials and studies recruiting in the UK
Cancer Chat forum
Talk to other people affected by cancer
Nurse helpline 0808 800 4040
Questions about cancer? Call freephone 9 to 5 Monday to Friday or email us
AACR Annual Meeting News: Read the latest session previews and recaps from the official news website.
Select "Patients / Caregivers / Public" or "Researchers / Professionals" to filter your results. To further refine your search, toggle appropriate sections on or off.
Home > About the AACR > History of the AACR > 115th Anniversary of the AACR > Landmarks in Cancer Research: 1907-1960
- AACR 115th Anniversary Event
- Landmarks in Cancer Research: Introduction
Landmarks in Cancer Research: 1907-1960
- Landmarks in Cancer Research: 1961-1990
- Landmarks in Cancer Research: 1991-2010
- Landmarks in Cancer Research: 2011-Present

- Sunlight exposure is linked to skin cancer. The first epidemiologic study of sunlight and skin cancer was reported; earlier observations had linked chronic skin conditions common in sailors to exposure to the radiation effects of the sun. Later work in animal models confirmed that skin cancer could be induced by ultraviolet light and sunlight. (1)(2)(3)
- American Association for Cancer Research is founded by four surgeons, five pathologists, and two biochemists ( right ) on May 7 in Washington, DC.
- Japanese cancer journal, Gann: Japanese Journal of Cancer Research (now titled Cancer Science ), is first published.
- Nine research papers are presented at the first Annual Meeting of the AACR in New York City.
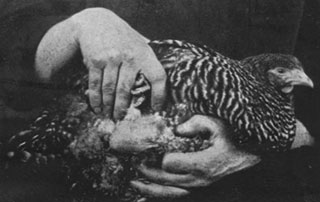
- Cell-free extracts transmit cancer from one animal to another. Cell-free agents were shown to transmit leukosis, a form of leukemia and lymphoma, and sarcomas in chickens. This finding would later be verified as evidence for viral initiation of cancer. (4)(5)

- Martha Tracy ( right ), from Women’s Medical College in Philadelphia (later dean of that college), becomes the AACR’s first female member.
- AACR writes President William H. Taft advocating funding for cancer research.
- Procedures for in vitro tissue culture are developed. The fundamental culture techniques, now ubiquitous in the laboratory, allowed researchers to study the evolution of tumor tissue under known conditions and to observe living cancer cells at every stage of growth. (6)
- French journal, Bulletin de l’Association Française pour l’Étude du Cancer , and the Italian journal, Tumori , are first published.

- AACR member Thomas S. Cullen, MD, presents “Education of the People as to What Can Be Done in Early Cases of Cancer” at the Annual Meeting. This appeal for public education led the Ladies’ Home Journal to publish “What Can We Do About Cancer,” the first consumer-oriented article about cancer.
- A group of volunteers—including AACR founding member and past president James Ewing—establishes the American Society for the Control of Cancer ( right ), precursor to the American Cancer Society.
- Alterations in chromosomes are postulated to cause tumor growth. From earlier work on sea urchin eggs and association of inappropriate segregation of chromosomes and changes in cell growth characteristics came the hypothesis that cancer was caused by abnormal chromosomes. (7)
- First experimental animal model of chemically induced cancer is developed. Repeated tarring of rabbit skin caused tumors. The discovery added to early evidence for the theory of chemical carcinogenesis, building upon the observation in 1775 of scrotal cancer in chimney sweeps. Later work published in the AACR’s The Journal of Cancer Research would isolate and identify the specific components of coal tar responsible. (8)(9)

- AACR begins publishing The Journal of Cancer Research , the first English language cancer journal.
- Oophorectomy decreases breast cancer in mice. Removal of the ovaries from female mice of a strain with a high incidence of spontaneous breast cancer resulted in a decrease in tumors. Later work published in The Journal of Cancer Research involving transplantation of ovaries into male mice showed an induction of mammary tumors, supporting the suggestion that hormones from the ovary could promote breast tumors. (10)(11)
- American Society for the Control of Cancer creates the first National Cancer Week as an extensive public education campaign.

- U.S. Public Health Service opens Office of Cancer Investigations at Harvard Medical School.
- Metabolic studies show that tumors exhibit anaerobic respiration. Whereas normal tissues use oxygen to break down nutrients for growth as their primary mode of respiration, it was observed that within tumors, cells respire anaerobically, fermenting sugars without oxygen. It will take several decades before hypoxia is revisited as a marker for tumors. (12)(13)
- Cancer is named one of the top three causes of death in America by U.S. Census Bureau.
- Genetic mutation is proposed as the origin of cancer. As an alternative to the infection theory of cancer, popular at the time because of the expansion of microbiology as a field of study, came the proposal that somatic mutation was the cause of cancer. As Mendel’s works were rediscovered in 1928, the field of genetics grew. The term “somatic mutation” had been coined in 1916. (14)(15)
- Cervical cancer cells are visible in smears of exfoliated vaginal cells. Findings of cervical cancer cells in smears were met with skepticism, and it would take until the 1960s before the “Pap” smear would become widely accepted as an effective method of screening and cancer prevention. (16)(17)
- X-rays are shown to be mutagenic. X-rays were shown to be mutagenic in the common fruit fly. This discovery formed the basis for thinking about how carcinogens participate in tumorigenesis. (18)
- First pure carcinogen, benzopyrene, is isolated from coal tar. The known cancer-causing environmental substance, coal tar, was fractionated into components and assayed in mouse models to identify the individual chemicals responsible for carcinogenesis. (19)
- Ransdell Act establishes the National Institute of Health.

- The American Journal of Cancer replaces The Journal of Cancer Research as the official AACR publication.
- Injected synthetic hormones induce breast cancer in mice. Building on work on endogenous hormones, it was demonstrated that addition of synthetic exogenous hormones such as folliculin (and in 1952, diethylstilbestrol) can induce cancer. (20)(21)
- Electron microscope is invented. The electron microscope permitted the visualization of minute subcellular structures, allowing observation of detailed differences between malignant and normal tissues. (22)

- National Cancer Institute Act establishes the National Cancer Institute (NCI) as an independent research institution.
- Transplantation of a single leukemic cell transmits leukemia in mice. Studies published in AACR’s The American Journal of Cancer showed that not all cancer cells behaved in an identical manner; some were uniquely capable of initiating and maintaining a tumor. This work laid the foundation for the later search for a cancer stem cell. (23)
- Telomeres are identified. The ends of chromosomes were shown to be protected by a structure that prevented their fusion. Later, it was shown that telomeres are repeated simple sequence elements that are added by an enzyme, telomerase, which is not normally expressed in somatic cells. In each cell division, telomeres shorten. When they become sufficiently truncated they cause the cells to enter into senescence and die, limiting the number of divisions a cell can undergo and suppressing tumor development. (24)(25)(26)
- Discovery of antigens explains why tumors can be transplanted within inbred strains. Previous work to transplant tumors had been successful in some instances but failed in others. The discovery of major histocompatibility antigens later led to an immunologic explanation that applied to grafts of normal tissue as well as to malignant tissue. (27)(28)
- Chemicals induce cancer in two distinct steps of initiation and promotion. Tumorigenesis was identified as a multistage disease, and it was shown that chemicals induce cancer in two distinct steps of initiation and promotion. A nonspecific irritant (wounding) was shown to promote tumorigenesis after initiation with a suboptimal dose of carcinogen (tarring or application of Shope papillomavirus to rabbit ears). Further study of the significance of cocarcinogenic action was later published in Cancer Research . (29)(30)
- Transplanted animal tumors are shown to grow blood vessels. Tumors transplanted into the ears of rabbits elicited a vascular network. This was early evidence of the phenomenon of angiogenesis, or new blood vessel growth, which would later become a target for antiangiogenesis cancer therapies. (31)
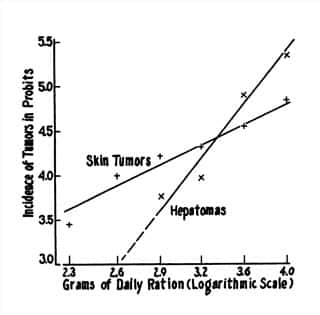
- Caloric restriction reduces tumors in mice. Studies published in The American Journal of Cancer and later in Cancer Research showed that caloric intake was proportional to the incidence of tumors of several kinds, including spontaneous mammary carcinomas and hepatomas in susceptible mouse strains and benzopyrene-induced skin tumors. Only recently, with the increasing prevalence of obesity in the global population, have the implications of the work been revisited. (32,33)

- Cancer Research replaces The American Journal of Cancer as AACR’s official journal.
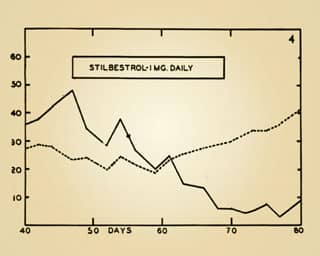
- Hormone dependence of prostate cancer is demonstrated. In a study published in Cancer Research , the therapeutic use of physical castration or chemical castration by treatment with estrogens was shown to decrease disease burden in metastatic prostate cancer whereas injection of androgens increased metastases. (34)
- DNA is identified as the active material in the genes of bacteria. It was not known whether the protein or DNA components of the chromosomes contained the information necessary for inheritance. This work showed that DNA contained the heritable information and set the stage for many important works and techniques. (35)
- American Society for the Control of Cancer becomes the American Cancer Society.
- Public Health Services Act designates NCI as a division of the National Institutes of Health (NIH).
- Atomic Bomb Casualty Commission is established to monitor the effects of radiation exposure.

- Nitrogen mustard is established as the first chemotherapeutic agent. Observational reports that soldiers exposed to nitrogen mustard during wartime had low white blood cell counts led to testing of nitrogen mustard as chemotherapy for cancer. Intravenous nitrogen mustard was shown to slow the growth of lymphomas and leukemias in patients refractory to radiation therapy, and it achieved remissions of a few months. Nitrogen mustard was approved for cancer treatment in 1949. (36)
- Nuremberg Code establishes the legal principle of voluntary consent for human subjects of research.
- At the 38th AACR Annual Meeting, May 16-17, a policy presentation titled, “On the Organization and Support of Cancer Research,” concludes that the AACR should advocate for increased funding for cancer research.
- First successful chemotherapy for childhood leukemia is reported. A synthetic folate antagonist achieved a three-month remission in 10 of 16 children with leukemia. Although not successful by today’s standards, this was an important result that would lead to further work on antimetabolites and the first generation of effective chemotherapeutic agents. (37)
- United Nations establishes the World Health Organization.
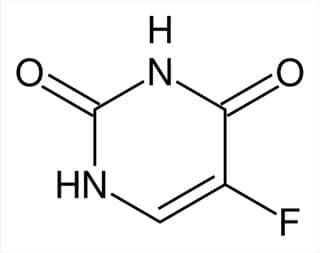
- First rationally conceived nucleotide analog chemotherapeutic agents are developed. Drug design had been primarily by trial and error. The design of molecules similar to the bases of DNA, but sufficiently different to prevent replication, proved an effective drug targeting approach that led to several chemotherapeutic drugs for cancer such as 6-mercaptopurine and 5-fluorouracil, which are still in use today. (38,39)
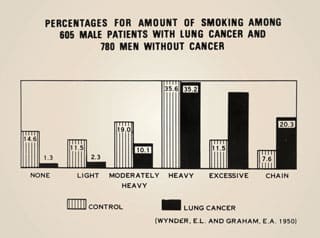
- Epidemiologic work links tobacco smoking to lung cancer. A retrospective analysis of the smoking habits of patients with lung cancer showed an association with tobacco. This was followed by a prospective study of male doctors that showed a clear relationship between smoking and lung cancer deaths. Tobacco exposure is now a known risk factor for many cancer types, accounting for an estimated 30% of all cancer mortality. (40-42)
- Leukemia in mice is shown to be transmissible by a virus. Leukemia had been considered an inherited disease before it was shown that it could be transmitted from one mouse strain to another by a virus and then passed from one generation to another via vertical transmission. These findings laid the groundwork for later research on other mouse tumor viruses and those in other species. (43)
- Cobalt-60 irradiator is developed. Radiotherapy previously had been carried out using radium, which was in limited supply and needed to be used in close proximity to the tumor. Radioactive cobalt provided a continuous source with greater ability to treat internal tumors, with less damage to the intervening tissue. Clinical cobalt-60 is still used in much of the developing world. (44)

- Ultrasound imaging is developed for detecting tumors. Although earlier studies had used ultrasound as a therapy and had examined its use as an imaging tool, research showed that ultrasound could detect differences in density between malignant and normal tissues. (45)

- AACR Annual Meeting abstracts are published for the first time as Proceedings of the American Association for Cancer Research (154 abstracts).
- Structure of DNA is described. Not only was the global structure of DNA identified, but how the bases pair and possible implications for methods of replication were also elucidated. (46)
- Human carcinoma cell line, HeLa, is established from the cells of Ms. Henrietta Lacks. The HeLa epithelial cell line is readily grown in laboratories worldwide and has become a fundamental tool for studying many aspects of molecular biology. Stable cell lines such as HeLa allow researchers to use genetically identical cells for experiments over long-term courses of repeated culturing in a manner not possible with primary cells. (47)
- Medical linear accelerator is developed for radiotherapy. Unlike early radiotherapy machines that used a radioactive source to generate X-rays, the linear accelerator produces a beam of electrons. This eliminated the need to replace the radioactive source and is limited in power by the length of the accelerator tube. (48)
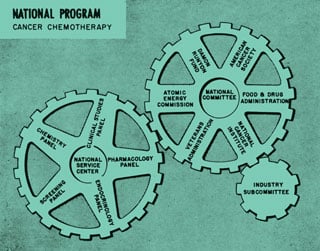
- Tumor clonogenic assay is developed. Although human cells had been cultured before, these new methods allowed cultures to be propagated from single human cells, enabling the kind of detailed genetic studies previously only possible for bacterial cells. (49)
- U.S. Congress funds National Chemotherapy Program to test compounds that might be effective against cancer.
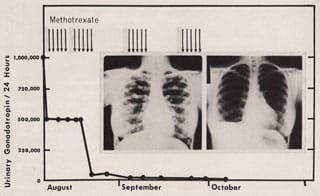
- First successful chemotherapy for solid tumors is reported. Building on earlier work on folate and aminopterin, another antifolate, methotrexate, was developed.The drug was shown to be effective in a small group of three patients with metastatic choriocarcinoma and chorioadenoma. (50)

- Elizabeth C. Miller ( right ) is the first woman elected to the AACR Board of Directors.
- Association of American Cancer Institutes (AACI) is founded. Its mission is to reduce the burden of cancer by enhancing the impact of North America’s leading academic cancer centers.
- Food Additives Amendment prohibits food additives shown to induce cancer in humans or animals.
- AACR membership passes 1,000.
- In vitro viral carcinogenesis is demonstrated. Earlier work had shown that viruses could be used to transmit cancer from one organism to another. New studies showed that chick embryo cells infected with Rous sarcoma virus continued to grow in culture and produce more virus. The infected cells had changes in morphology and rapid, disordered growth characteristic of cancer cells. (51)

- DNA repair after radiation is demonstrated. Chinese hamster ovary cells subjected to X-irradiation and surviving did not display heritable damage but repaired the damage prior to cell division. This finding confirmed the presence of DNA repair mechanisms, later shown to be defective in some cancers. (52)
- Dose-response relationship is shown in radiation-induced leukemia. Radiation carcinogenesis was unequivocally established in human populations, and the nature of the dose-response relationship was described. (53)
- Radioimmunoassay is developed. The radioimmunoassay uses antibodies to detect the amounts of specific proteins in a solution. Originally developed to measure insulin levels in the blood of diabetics, this technique is now the basis for diagnostic tests to measure serum proteins and biomarkers, such as prostate-specific antigen, although now the detection mechanism often uses fluorescent rather than radioactive labeling. (54)
- American Cancer Society urges widespread use of Pap smear to detect cervical and uterine cancers.
- Philadelphia chromosome is discovered . An abnormally small chromosome was identified in the neoplastic cells of patients with chronic myelogenous leukemia. This small chromosome, later named the Philadelphia chromosome after the city in which it was discovered, was the first chromosomal abnormality found to be consistently associated with a specific human neoplasm. (55)
- Growth factors are purified and identified. The fact that growth factors were necessary for cells to survive and replicate had long been known, but the individual components of serum responsible had not been identified. The purification of nerve-growth factor led to the identification of other growth factors, their cognate receptors, and their complex signaling pathways. These pathways have emerged as novel targets for therapies such as those targeting the epidermal growth factor receptor (EGFR). (56)
- Screening techniques for prevention of colon cancer are adopted. The sigmoidoscope permitted early identification of colorectal cancer as well as precancerous polyps, leading to increased survival rates. It is estimated that screening by sigmoidoscopy, colonoscopy, barium enema, or fecal occult blood testing may result in a 20% decrease in colorectal cancer mortality. (57,58)
- Dubreuilh W. Epitheliomatose d’origine solaire. Ann Dermatol Syphiligr 1907;8:387–416.
- Findlay GM. Ultra-violet light and skin cancer. Lancet 1928;212:1070–3.
- Roffo AH. Krebs und sarkom durch ultraviolett- und sonnenstrahlen. Z Krebsforsch 1935;41:448–67.
- Ellermann V, Bang O. Experimentelle leukamie bei huhnern. Erscheinungsort nicht ermittelbar. Vorlaufige Mitteilung Centralbl f Bakteriol 1908; xlvi, 4.
- Rous P. A transmissible avian neoplasm (sarcoma of the common fowl). J Exp Med 1910;12:696–705.
- Carrel A, Burrows MT. Cultivation of sarcoma outside of the body: a second note. JAMA 1910;55:1554.
- Boveri T. Zur frage der entstehung maligner tumoren. Jena, Germany: Gustav Fisher; 1914.
- Yamagiwa K, Ichikawa K. [Repeated painting of coal tar onto rabbits’ ears causes carcinomas]. J Imperial Univ Tokyo 1915;15:295−344.
- Yamagiwa K, Ichikawa K. Experimental study of the pathogenesis of carcinoma. CA Cancer J Clin 1977;27:174−81.
- Lathrop AE, Loeb L. Further investigations on the origin of tumors in mice. III. On the part played by internal secretion in the spontaneous development of tumors. Cancer Res 1916;1:1−19.
- Murray WS. Ovarian secretion and tumor incidence. Cancer Res 1928;12:18−25.
- Warburg O, Posener K, Negelein E. [VII. The metabolism of the cancer cell]. Biochem Z 1924;152:319−44.
- Weinhouse S. On respiratory impairment in cancer cells. Science 1956;124:267−9.
- Bauer KH. Mutationstheorie der geschwulst-entstehung. Berlin, Germany: Springer; 1928.
- Tyzzer EE. Tumor immunity. Cancer Res 1916;1:125−56.
- Papanicolaou GN. New cancer diagnosis. CA Cancer J Clin 1973;23:174−9.
- Papanicolaou GN. A survey of the actualities and potentialities of exfoliative cytology in cancer diagnosis. Ann Intern Med 1949;31:661−74.
- Muller HJ. The production of mutations by X-rays. Proc Natl Acad Sci U S A 1928;14:714−26.
- Kennaway EL. Further experiments on cancer-producing substances. Biochem J 1930;24:497−504.
- Lacassagne MA. [Appearance of mammary cancers in male mice subjected to folliculin injections]. Comptes Rendus de l’Academie des Sciences 1932;195:630−2.
- Dunning WF, Curtis MR. The incidence of diethylstilbestrol-induced cancer in reciprocal F hybrids obtained from crosses between rats of inbred lines that are susceptible and resistant to the induction of mammary cancer by this agent. Cancer Res 1952;12:702−6.
- Knoll M, Ruska E. Das elektronenmikroskop. Zeitschrift für Physik 1932;78:318−39.
- Furth J, Kahn MC, Breedis C. The transmission of leukemia of mice with a single cell. Am J Cancer 1937;31:276−82.
- McClintock B. The fusion of broken ends of sister half-chromatids following chromatid breakage at meiotic anaphases [dissertation]. Columbia, MO: University of Missouri; 1938.
- McClintock B. The discovery and characterization of transposable elements: the collected papers of Barbara McClintock. New York: Garland Publishing; 1987.
- Müller HJ. The remaking of chromosomes. Collecting Net 1938;13:181−98.
- Gorer PA. The genetic and antigenic basis of tumour transplantation. J Pathol Bacteriol 1937;44:691−7.
- Gorer PA, Lyman S, Snell GD. Studies on the genetic and antigenic basis of tumour transplantation linkage between a histocompatibility gene and “fused” in mice. Proc R Soc Lond B Biol Sci 1948;135:499−505.
- Kidd JG, Rous P. The carcinogenic effect of a papilloma virus on the tarred skin of rabbits: II. Major factors determining the phenomenon: the manifold effects of tarring. J Exp Med 1938;68:529−62.
- Berenblum I. The mechanism of carcinogenesis. A study of the significance of cocarcinogenic action and related phenomena. Cancer Res 1941;1:807−14.
- Ide AG, Baker NH, Warren SL. Vascularization of the brown Pearce rabbit epithelioma transplant as seen in the transparent ear chamber. Am J Roentgenol 1939;42:891−9.
- Tannenbaum A. The initiation and growth of tumors. Introduction. I. Effects of underfeeding. Am J Cancer 1940;38:335−50.
- Tannenbaum A, Silverstone H. The influence of the degree of caloric restriction on the formation of skin tumors and hepatomas in mice. Cancer Res 1949;9:724−7.
- Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Res 1941;1:293−7.
- Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a deoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 1944;79:137−58.
- Goodman LS, Wintrobe MM, Dameshek W, Goodman MJ, Gilman A, McLennan MT. Nitrogen mustard therapy. Use of methyl-bis(beta-chloroethyl)amine hydrochloride and tris(beta-chloroethyl)amine hydrochloride for Hodgkin’s disease, lymphosarcoma, leukemia and certain allied and miscellaneous disorders. JAMA 1946;132:126−32.
- Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med 1948;238:787−93.
- Hitchings GH, Elion GB, Falco EA, Russell PB, Sherwood MB, Vanderwerff H. Antagonists of nucleic acid derivatives: I. The Lactobacillus casei model. J Biol Chem 1950;183:1−9.
- Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, et al. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature 1957;179:663−6.
- Doll R, Hill AB. Smoking and carcinoma of the lung; preliminary report. Br Med J 1950;2:739−48.
- Doll R, Hill AB. The mortality of doctors in relation to their smoking habits; a preliminary report. Br Med J 1954;1:1451−5.
- Doll R, Hill AB. Lung cancer and other causes of death in relation to smoking; a second report on the mortality of British doctors. Br Med J 1956;2:1071−81.
- Eddy BE, Stewart SE, Berkeley W. Cytopathogenicity in tissue culture by a tumor virus from mice. Proc Soc Exp Biol Med 1958;98:848−51.
- Johns HE, Bates LM, Epp ER, Cormack DV, Fedorux SO, Morrison A, et al. 1,000-curie cobalt 60 units for radiation therapy. Nature 1951;168:1035−6.
- French LA, Wild JJ, Neal D. Detection of cerebral tumors by ultrasonic pulses; pilot studies on postmortem material. Cancer 1950;3:705−8.
- Watson JD, Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 1953;171:737−8.
- Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med 1953;97:695−710.
- Miller CW. Travelling-wave linear accelerator for X-ray therapy. Nature 1953;171:297−8.
- Puck TT, Marcus PI. A rapid method for viable cell titration and clone production with HeLa cells in tissue culture: the use of X-irradiated cells to supply conditioning factors. Proc Natl Acad Sci U S A 1955;41:432−7.
- Hertz R, Li MC, Spencer DB. Effect of methotrexate therapy upon choriocarcinoma and chorioadenoma. Proc Soc Exp Biol Med 1956;93:361−6.
- Vigier P, Golde A. Growth curve of Rous sarcoma virus on chick embryo cells in vitro. Virology 1959;8:60−79.
- Elkind MM, Sutton H. X-ray damage and recovery in mammalian cells in culture. Nature 1959;184:1293−5.
- Armitage P, Court Brown WM, Doll R, Mewissen DJ. Dose-response relationship in radiation leukaemia. Nature 1959;184:1669−70.
- Yalow RS, Berson SA. Assay of plasma insulin in human subjects by immunological methods. Nature 1959;184:1648−9.
- Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science 1960;132:1497.
- Cohen S. Purification of a nerve-growth promoting protein from the mouse salivary gland and its neuro-cytotoxic antiserum. Proc Natl Acad Sci U S A 1960;46:302−11.
- Cameron AB, Thabet RJ. Sigmoidoscopy as part of routine cancer clinic examinations with correlated fecal chemistry and colon cytologic studies. Surgery 1960;48:344−50.
- Vogelaar I, van Ballegooijen M, Schrag D, Boer R, Winawer SJ, Habbema JD, et al. How much can current interventions reduce colorectal cancer mortality in the U.S.? Mortality projections for scenarios of risk-factor modification, screening, and treatment. Cancer 2006;107:1624−33.
Your donation to the American Association for Cancer Research helps our more than 58,000 members worldwide drive progress against cancer.

- History of the AACR
- The Past, Present, and Future of HPV: Can...
Advertisement

The Hallmarks of Precancer
M.M. Stangis, Z. Chen, J. Min, S.E. Glass, J.O. Jackson, and M.D. Radyk contributed equally to this article.
Cancer Discov 2024;14:683–9
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Get Permissions
- Cite Icon Cite
- Search Site
- Version of Record April 4 2024
Mary M. Stangis , Zhengyi Chen , Jimin Min , Sarah E. Glass , Jordan O. Jackson , Megan D. Radyk , Xen Ping Hoi , W. Nathaniel Brennen , Ming Yu , Huy Q. Dinh , Robert J. Coffey , Martha J. Shrubsole , Keith S. Chan , William M. Grady , Srinivasan Yegnasubramanian , Costas A. Lyssiotis , Anirban Maitra , Richard B. Halberg , Neelendu Dey , Ken S. Lau; The Hallmarks of Precancer. Cancer Discov 1 April 2024; 14 (4): 683–689. https://doi.org/10.1158/2159-8290.CD-23-1550
Download citation file:
- Ris (Zotero)
- Reference Manager
Research on precancers, as defined as at-risk tissues and early lesions, is of high significance given the effectiveness of early intervention. We discuss the need for risk stratification to prevent overtreatment, an emphasis on the role of genetic and epigenetic aging when considering risk, and the importance of integrating macroenvironmental risk factors with molecules and cells in lesions and at-risk normal tissues for developing effective intervention and health policy strategies.
Client Account
Citing articles via, email alerts.
- Online First
- Online ISSN 2159-8290
- Print ISSN 2159-8274
AACR Journals
- Blood Cancer Discovery
- Cancer Discovery
- Cancer Epidemiology, Biomarkers & Prevention
- Cancer Immunology Research
- Cancer Prevention Research
- Cancer Research
- Cancer Research Communications
- Clinical Cancer Research
- Molecular Cancer Research
- Molecular Cancer Therapeutics
- Info for Advertisers
- Information for Institutions/Librarians
- Privacy Policy
- Copyright © 2023 by the American Association for Cancer Research.
This Feature Is Available To Subscribers Only
Sign In or Create an Account
Cancer patients can now be 'matched' to best treatment with DNA and lab-dish experiments
Identifying the most effective cancer treatment for a given patient from the get-go can help improve outcomes.
Despite many efforts to find better, more effective ways to treat cancer, it remains a leading cause of death by disease among children in the U.S.
Cancer patients are also getting younger. Cancer diagnoses among those under 50 has risen by about 80% worldwide over the past 30 years. As of 2023, cancer is the second-leading cause of death both in the U.S. and around the world. While death rates from cancer have decreased over the past few decades, about 1 in 3 patients in the U.S. and 1 in 2 patients worldwide still die from cancer.
Despite advances in standard cancer treatments, many cancer patients still face uncertain outcomes when these treatments prove ineffective. Depending on the stage and location of the cancer and the patient's medical history, most cancer types are treated with a mix of radiation, surgery and drugs. But if those standard treatments fail, patients and doctors enter a trial-and-error maze where effective treatments become difficult to predict because of limited information on the patient's cancer.
My mission as a cancer researcher is to build a personalized guide of the most effective drugs for every cancer patient. My team and I do this by testing different medications on a patient's own cancer cells before administering treatment, tailoring therapies that are most likely to selectively kill tumors while minimizing toxic effects.
In our newly published results of the first clinical trial combining drug sensitivity testing with DNA testing to identify effective treatments in children with cancer, an approach called functional precision medicine , we found this approach can help match patients with more FDA-approved treatment options and significantly improve outcomes.
What is functional precision medicine?
Even though two people with the same cancer might get the same medicine, they can have very different outcomes. Because each patient's tumor is unique, it can be challenging to know which treatment works best.
To solve this problem, doctors analyze DNA mutations in the patient's tumor, blood or saliva to match cancer medicines to patients. This approach is called precision medicine . However, the relationship between cancer DNA and how effective medicines will be against them is very complex. Matching medications to patients based on a single mutation overlooks other genetic and nongenetic mechanisms that influence how cells respond to drugs.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
How to best match medicines to patients through DNA is still a major challenge. Overall, only 10% of cancer patients experience a clinical benefit from treatments matched to tumor DNA mutations.
Functional precision medicine takes a different approach to personalizing treatments. My team and I take a sample of a patient's cancer cells from a biopsy, grow the cells in the lab and expose them to over 100 drugs approved by the Food and Drug Administration. In this process, called drug sensitivity testing , we look for the medications that kill the cancer cells.
New clinical trial results
Providing functional precision medicine to cancer patients in real life is very challenging. Off-label use of drugs and financial restrictions are key barriers. The health of cancer patients can also deteriorate rapidly, and physicians may be hesitant to try new methods.
But this is starting to change. Two teams in Europe recently showed that functional precision medicine could match effective treatments to about 55% of adult patients with blood cancers such as leukemia and lymphoma that did not respond to standard treatments.
Most recently, my team's clinical trial focused on childhood cancer patients whose cancer came back or didn't respond to treatment. We applied our functional precision medicine approach to 25 patients with different types of cancer.
Our trial showed that we could provide treatment options for almost all patients in less than two weeks. My colleague Arlet Maria Acanda de la Rocha was instrumental in helping return drug sensitivity data to patients as fast as possible. We were able to provide test results within 10 days of receiving a sample, compared with the roughly 30 days that standard genomic testing results that focus on identifying specific cancer mutations typically take to process.
Most importantly, our study showed that 83% of cancer patients who received treatments guided by our approach had clinical benefit, including improved response and survival.
Expanding into the real world
Functional precision medicine opens new paths to understanding how cancer drugs can be better matched to patients. Although doctors can read any patient's DNA today, interpreting the results to understand how a patient will respond to cancer treatment is much more challenging. Combining drug sensitivity testing with DNA analysis can help personalize cancer treatments for each patient.
I, along with colleague Noah E. Berlow , have started to add artificial intelligence to our functional precision medicine program. AI enables us to analyze each patient's data to better match them with tailored treatments and drug combinations. AI also allows us to understand the complex relationships between DNA mutations within tumors and how different treatments will affect them.
— 'Very concerning': Microplastics can accumulate in cancer cells and may help them spread, study hints
— Gut bacteria linked to colorectal cancer in young people
— New mRNA 'cancer vaccine' trial launches in UK
My team and I have started two clinical trials to expand the results of our previous studies on providing treatment recommendations through functional precision medicine. We're recruiting a larger cohort of adults and children with cancers that have come back or are resistant to treatment.
The more data we have, the easier it will become to understand how to best treat cancer and ultimately help more patients access personalized cancer treatments.
This edited article is republished from The Conversation under a Creative Commons license. Read the original article .

Diana Azzam is an Assistant Professor and Research Director of the newly established Center for Advancing Personalized Cancer Treatments (CAPCT) at Florida International University. She has a Masters in Biochemistry from the American University of Beirut, Lebanon and a PhD in Biochemistry & Molecular Biology from the University of Miami, Florida. Her lab focuses on implementing functional precision medicine (FPM) approaches in adult and pediatric cancer patients that have run out of treatment options. Working with local hospitals including Nicklaus Children's Hospital and Cleveland Clinic Florida, her lab delivers individualized treatment plans based on a patient's cancer genomic profile and ex vivo drug response. She is currently engaged in two clinical studies to assess feasibility and clinical utility of FPM in relapsed/refractory patients with childhood cancer (ClinicalTrials.gov registration: NCT05857969) and adult cancer (ClinicalTrials.gov registration: NCT06024603). She is working on setting up the first CLIA-certified lab in the State of Florida dedicated for functional cancer drug testing. Her goal is to launch large-scale prospective multi-center randomized clinical trials to better assess efficacy of FPM approaches in the treatment of refractory/relapsed cancers. In parallel, she is working on utilizing FPM as a tool to reduce health disparities in childhood cancer patients from minority populations. She is also integrating a novel machine learning approach to identify specific biomarkers among minority populations that can be targeted using FDA-approved drugs. Her lab also investigates cancer stem cells and how they may result from chronic environmental exposures to toxic metals such as arsenic.
Catherine, Princess of Wales, announces cancer diagnosis
'Very concerning': Microplastics can accumulate in cancer cells and may help them spread, study hints
Gravitational waves reveal 1st-of-its-kind merger between neutron star and mystery object
Most Popular
- 2 Viking Age women with cone-shaped skulls likely learned head-binding practice from far-flung region
- 3 Largest 3D map of our universe could 'turn cosmology upside down'
- 4 Mass die-off half a billion years ago caused by shifting tectonic plates, ancient rocks reveal
- 5 Prehistoric henge accidentally discovered in England in search for Anglo-Saxon hermit
- 2 Why did Europe's hunter-gatherers disappear?
- 3 Ancient Indigenous lineage of Blackfoot Confederacy goes back 18,000 years to last ice age, DNA reveals
Terry Fox: A Hero's Journey of Hope and Determination Daily Sports History
Join us as we celebrate the extraordinary life and legacy of Terry Fox, a Canadian icon whose courageous Marathon of Hope inspired millions around the world. In this short episode, we delve into Fox's remarkable journey—a marathon across Canada to raise funds for cancer research, despite battling cancer himself.Discover the indomitable spirit of Terry Fox as he ran over 5,000 kilometers on his prosthetic leg, capturing the hearts of Canadians and people worldwide. Hear stories of perseverance, resilience, and the power of one person to make a difference in the fight against cancer.Through concise storytelling, we honor Terry Fox's enduring legacy as a beacon of hope, courage, and determination, inspiring generations to continue the fight against cancer and pursue their dreams against all odds. We are being featured on PODCAST GURU: https://app.podcastguru.io/podcast/daily-sports-history-1715849627 Website: dailysportshistory.com Email: [email protected] YouTube: YouTube.com/@dailysportshistory Twitter: twitter.com/dailysportshis Facebook: facebook.com/profile.php?id=61551687917253&mibextid=ZbWKwL Tiktok: tiktok.com/@daily.sports.history?_t=8hHPnNSCqfm&_r=1 #sports #sportshistory #sportspodcast #podcast #TerryFox #MarathonOfHope #CancerResearch #InspirationalFigures #CanadianHeroes #Activism #CancerAwareness #Humanitarianism
- Episode Website
- More Episodes
- Copyright Ethan Reese
Top Podcasts In Sports
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Cold Spring Harb Perspect Biol
- v.13(6); 2021 Jun
A History of Cancer Research: Tumor Viruses
Early studies of transmissible tumors in chickens provided evidence that viruses such as avian leukosis virus (ALV) and Rous sarcoma virus (RSV) can cause cancer in these animals. Doubts about the relevance to human tumors and failures to replicate some early work meant the field of tumor virology followed a bumpy course. Nevertheless, viruses that can cause cancers in rodents and humans were ultimately identified, and several Nobel prizes were awarded for work in this area. In this excerpt from his forthcoming book on the history of cancer research, Joe Lipsick looks back at the early history of tumor virus research, from some of the early false starts and debates, to discovery of reverse transcriptase, and identification of human papilloma virus (HPV) as the major cause of cervical cancer.
HERE A CHICK, THERE A CHICK
The early twentieth century witnessed the rise and ignoble fall of Fibiger's Nobel Prize–winning “discovery” of worms as a cause of cancer. But all was not rotten in the state of Denmark. Elsewhere at the University of Copenhagen, Vilhelm Ellerman and Oluf Bang were also testing whether cancer might be an infectious disease. In 1908 they published a paper entitled “Experimental Leukemia in Chickens.” They had found that leukemia, a cancer of the blood cells, could be transmitted from one bird to another by injection ( Fig. 1 ). Furthermore, the causative agent was able to pass through filters too fine to permit passage of any cells, animal or bacterial. Such filterable agents, first discovered in plants, eventually became known as viruses. Ellermann and Bang had discovered avian leukosis virus (ALV), the first known tumor virus. For reasons that remain unclear, their work did not attract the attention it deserved. One criticism was that although one could transmit this leukemia by injection, there was no observable transmission from bird to bird in the absence of injection. Furthermore, not every injected animal developed the disease (∼40%), and those that did often took a while to do so (6–12 mo). There was also a concern that the increased number of white blood cells might be a physiological response to infection, rather than a true malignant proliferation. Finally, it was not then widely accepted that animals, particularly non-mammals, were good models for human disease.

Ellermann and Bang's drawings of normal chicken bone marrow ( left ) and leukemic chicken bone marrow ( right ). The normal marrow contains dense, dark trabeculae of bone. The cells in the spaces between trabeculae are nucleated erythrocytes (red blood cells). The leukemic marrow contains little bone, many immature leukocytes (white blood cells, later shown to be B lymphocytes), and very few erythrocytes. (Reprinted from Ellermann V, Bang O. 1909. Z Hygeine Infektionskrakheiten 63: 231–273.)
Two years after Ellermann and Bang had published their work, Peyton Rous at the Rockefeller Institute described a transmissible sarcoma in chickens ( Fig. 2 ). The original tumor “was found in a barred Plymouth Rock hen of light color and pure blood” that was brought to him by a chicken breeder. Rous minced the tumor into small pieces, injected part into the other breast of the same chicken and part into two other chickens of the same brood. The original chicken died of widespread cancer 35 d later. By this time, one of the other injected chickens had also developed a palpable tumor. In his initial report Rous noted, “The tumor is at best so difficult of propagation that no attempts have been made to determine whether it can be transmitted by cell-fragments, or by cell-free derivatives.”

The origins of Rous sarcoma virus. ( Top ) A sarcoma caused by injection of fragments of the transmissible tumor. ( Bottom ) Histopathologic evidence of a sarcoma invading into muscle. (Reprinted from Rous P. 1910. J Exp Med 12: 696–705.)
However, within a year Rous had found that by passage from chicken to chicken, the transmissible tumor had become increasingly aggressive and was now capable of metastasizing. By 1911 he was able to follow in the footsteps of Bang and Ellermann and transmit this cancer by a filterable agent. This virus eventually became known as Rous sarcoma virus (RSV). As before, the work was not generally accepted as proof that cancer could be caused by an infectious agent. On the contrary, physicians were spending considerable time and effort trying to disabuse the public of the view that human cancer was infectious. This mistaken belief often resulted in the shunning or even the quarantine of patients afflicted with cancer. In search of better career prospects, Rous stopped working on RSV a few short years after publishing his landmark paper in 1911.
Extracts of chicken tumors from Rous's laboratory did make it across the pond to England, only to become entangled in a rather Dickensian tale. It began with an unusual proposition made to a studious railway stationmaster named William Bullock. If Bullock would agree to take a wealthy but childless benefactor's name, he would be left a small fortune with which he could attend medical school. Thus, was he reborn as William Gye. He enrolled at Edinburgh University, pursued a career in cancer research, and eventually was able to repeat the experiments of Rous. Taking things one step further, he then found that he could amplify the infectious material in vitro using fragments of chicken embryos. He also claimed to have isolated similar infectious agents in tumors from mice, from rats, and from humans, all of which could cause tumors in chickens.
Gye then collaborated with J.E. Barnard, a wealthy hatter and amateur microscopist, to obtain what they believed were images of the infectious cancer virus particles. These studies were published in 1925 as back-to-back papers in The Lancet . Not surprisingly, this work attracted wide attention in the popular press, helping to again fuel fears about the infectious nature of human cancer. Ultimately, none of this work stood the test of time, except for Gye's replication of Rous's work on chicken viruses. Gye went on to become a very successful cancer research administrator, eventually serving as the Director of the Imperial Cancer Research Fund Laboratories at Mill Hill.
The field of tumor virology itself also followed a rather bumpy course. Although additional tumor viruses were isolated from chickens in Japan and elsewhere, critics harped on the lack of evidence for similar viruses in mammals. In 1933 Richard Shope at the Rockefeller Institute identified a virus capable of causing papillomas (warts) in rabbits. Rous himself propagated and studied this virus for many years. A few years later John Bittner discovered a transmissible mammary cancer in mice that was caused by a milk-borne virus that became known as mouse mammary tumor virus (MMTV). In the early 1950s, Ludwik Gross identified two more mouse tumor viruses, a murine leukemia virus and a polyoma virus, which caused many different types of cancer. Tumor viruses were then identified in a variety of other mammals, including rats, cats, cows, and monkeys.
An adenovirus isolated from human tissue was shown to cause cancer in rodents in 1962, but was ultimately found not to be a cause of human cancer. Simian vacuolating virus 40 (SV40), a contaminant discovered in cells used to produce polio vaccine, was also shown to cause cancer in rodents in 1962. However, there has been a lack of convincing evidence that SV40 causes human cancer, despite much effort.
Finally, in 1964 Michael Epstein and Yvonne Barr published evidence of the first human tumor virus. They had discovered a herpes-like virus in the lymphoblasts of patients with Burkitt's lymphoma, a cancer endemic in children in tropical Africa. Epstein–Barr virus was later shown to cause both mononucleosis (a benign proliferation of B lymphocytes) and a form of nasopharyngeal cancer endemic in certain regions of China. On the heels of the discovery of the first human tumor virus, Rous was finally awarded the Nobel Prize in Physiology or Medicine in 1966 for his discovery of a chicken sarcoma virus 55 years earlier.
THE HUMAN CONDITION
Rising political pressure in the 1970s caused President Richard Nixon to announce a “war on cancer.” Ironically, much of this pressure came from Mary Lasker, a champion of medical research whose husband Albert Lasker had created advertising campaigns that greatly increased the popularity of cigarettes ( Fig. 3 ). Viruses turned out to be important causes of cancer in domesticated animals, such as chickens, laboratory mice, house cats, and cattle. However, despite the expenditure of much effort and many dollars, viruses were not found to cause the majority of human cancers. There were a number of highly publicized false leads, derisively referred to as “rumor viruses.” These reports were most often a result of contamination of human cells by animal viruses in the laboratory. The Special Virus Cancer Program, a directed medical research effort within the National Cancer Institute (NCI), had been charged with the discovery of new human cancer viruses. The program began in 1964, grew to consume substantial resources, and was eventually discontinued in the late 1970s. In part this was due to criticism from scientists outside the walls of the NCI, who favored peer-reviewed research directed by independent individual investigators rather than a centralized bureaucracy that managed large research contracts.

The Laskers’ contributions to cancer dissemination ( top ) and to political pressure for increasing spending on research for a cancer cure ( bottom , advertisement in The Washington Post ). ( Top , From the collection of Stanford Research Into the Impact of Tobacco Advertising [tobacco.stanford.edu]; bottom , https://profiles.nlm.nih.gov/spotlight/tl/catalog/nlm:nlmuid-101584665X20-doc .)
Eventually other human tumor viruses were discovered. Most of these viruses cause relatively rare types of cancer (e.g., HTLV-I causes lymphoid cancers of the skin; HTLV-II causes a rare form of leukemia; HHV-8 causes Kaposi's sarcoma; MCV causes Merkel cell cancer). One notable exception is a family of human papilloma viruses (HPVs) very similar to those discovered in rabbits by Shope. Two such viruses (HPV16 and HPV18) were shown by Harald zur Hausen in the 1980s to be the major cause of cancer of the uterine cervix in women. HPV also causes head and neck cancers and anogenital cancers. These observations led to a Nobel Prize in Physiology or Medicine for zur Hausen in 2008, and to the creation of preventive HPV vaccines, the first of which was approved by the FDA for clinical use in 2006.
In addition to viruses that appear to directly cause human cancer, certain viruses and other infectious agents appear to cause cancer in part via the response of the host to infection. For example, infection with hepatitis B and C viruses is strongly associated with cancer of the liver, infection with Helicobacter pylori bacteria is strongly associated with cancer of the stomach, and infection with Schistosoma haematobium is strongly associated with bladder cancer in some countries. Hepatitis B virus and H. pylori have been shown to encode proteins that can promote cell proliferation. However, these cancers all appear to require repeated cycles of infection, chronic inflammation, and tissue repair, resulting in the continued proliferation of cells that have also been exposed to environmental carcinogens. The result is an unholy alliance of three old rivals—the irritation theory, the germ theory, and the mutagen theory of cancer.
THE RISE OF THE QUANTS
In the mid-twentieth century, a number of physicists turned their attention from physics to biology. A particularly influential book called What Is Life? by Erwin Schrödinger proposed that genetic information might be contained within a chemical form. This idea spurred a group of physicists-turned-biologists to focus their attention on simpler and simpler genetic systems that could be studied by quantitative methods. The result was the Phage Group led by Max Delbrück, Alfred Hershey, and Salvador Luria. By studying the viruses of bacteria (bacteriophage or phage), they and their colleagues were able to deduce many of the basic principles of molecular biology. A by-product of their efforts was the development of methods for quantitative virology, largely based on the plaque assay first described by Félix d'Hérelle in 1917.
Similar advances in animal virology required the development of methods for studying viruses in systems simpler than a whole animal. In the early twentieth century, Alexis Carrel, a Nobel Prize–winning French surgeon, developed and publicized rather complex and somewhat mystical methods for culturing fragments of tissue in the laboratory. In 1928 Carrel reported that he could use these cultures to propagate RSV. He also claimed to have kept a continuous culture of embryonic chicken heart cells alive for decades in his laboratory at the Rockefeller Institute. His work attracted the attention of Charles “Lucky Lindy” Lindbergh, who with Carrel sought a path to physical immortality ( Fig. 4 ). Eventually Carrel's immortal chicken heart experiments failed to be repeated by others. Most likely, new cells had been continually added via the embryo extracts used to “feed” the cultures. Carrel later returned to Europe, where he became an advocate of eugenics for the Vichy government in Nazi-occupied France, thus sharing sympathies with Lindbergh.

Charles Lindbergh, Alexis Carrel, and the quest for immortality. (Image from the National Portrait Gallery.)
The fields of animal cell culture and animal virology progressed slowly until the 1940s, when recurrent polio epidemics spurred intense interest in animal (including human) virology. In 1948, John Enders and his colleagues succeeded in propagating poliovirus in cultures of human embryonic tissue fragments. Their work led to the intensely competitive development of polio vaccines by Jonas Salk and Albert Sabin. Based on the work of Enders, in 1954 Renato Delbecco and Marguerite Vogt developed a plaque assay for poliovirus that was similar to the method used to study the viruses that had lysed bacteria. At the same time, Harry Eagle at the National Institutes of Health (NIH) was systematically determining the requirements for animal cell growth. The result was a defined medium that, when supplemented with relatively small amounts of animal serum, permitted the reproducible growth of animal cells in culture without the need for embryo extracts or plasma clots. Together these powerful methods were then applied to the quantitative study and isolation of mutants of an ever-increasing number of animal viruses, including other important human pathogens like influenza.
But what about tumor viruses? How could one study viruses that caused cells to proliferate rather than die? The key insight came from Howard Temin and Harry Rubin, a graduate student and a postdoctoral fellow working in Dulbecco's laboratory. They infected dishes of adherent fibroblasts from chicken embryos with different dilutions of RSV, layered agar over the cultures, and then watched and waited. Following infection with very dilute stocks of virus, distinct patches of transformed cells appeared ( Fig. 5 ). Normal fibroblasts are flat, spindly cells that stop proliferating once they touch one another (contact inhibition). By contrast, RSV-transformed fibroblasts round up and keep proliferating, eventually forming small mounds of cells (transformed foci) that are very refractile when seen through a phase-contrast microscope. The agar overlay was an important modification of the method described two years earlier by Manaker and Groupé, because it greatly decreased secondary foci caused by subsequent rounds of infection or by detachment and diffusion of transformed cells. Once again using the logic of the Phage School, Temin and Rubin were able to rapidly and readily quantitate stocks of a tumor virus in a far easier fashion than had been possible with assays in whole animals or chicken eggs. Temin and Rubin observed a linear relationship between the concentration of the virus and the number of foci that extended over a thousandfold range. These results implied that infection with a single viral particle was sufficient to cause the oncogenic transformation of a normal cell. Therefore, by isolating virus from a single focus formed at a low concentration of virus, one could isolate a biological clone that had arisen from a single virus particle.

Temin and Rubin's focus assay for morphologic transformation by Rous sarcoma virus. ( Left ) Experimental scheme. ( Center ) A transformed focus visualized by phase contrast microscopy. ( Right ) Relationship between viral concentration and number of foci. ( Center and right panels from Temin H, Rubin H. 1958. Virology 6: 669–688, with permission from Elsevier.)
PROVIRAL HERESY
Temin continued these studies in his own laboratory. He noticed that some isolates of RSV transformed cells with a cobblestone-like morphology (round), whereas other isolates of RSV transformed cells with a spindly morphology (fusiform) ( Fig. 6 ). Remarkably, these characteristics bred true. Cells transformed by a “round” variant of RSV and the progeny of these cells remained “round,” as did naive cells transformed by viruses isolated from “round” cells. Similarly, cells transformed by the “fusiform” variant of RSV and the progeny of those cells remained “fusiform,” as did naive cells transformed by viruses isolated from “fusiform” cells. Furthermore, the conversion of one viral variant into another occurred very rarely, if ever. These observations led Temin to propose that the transformed state was a stably inherited property of the infected cells. To account for this heritable state, he further proposed that the genetic material of the virus somehow became part of the genetic material of the cell.

Cells transformed by fusiform (f) variant and round (r) morphological variants of Rous sarcoma virus. (Photomicrograph by Peter Vogt from Coffin J, et al. 1997. Retroviruses . Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, with permission from Peter Vogt.)
There were precedents for viral integration into the host genome in the world of bacteriophage biology. In the 1920s, Eugene and Elizabeth Wollman at the Institut Pasteur in Paris had discovered a latent form of bacteriophage that did not cause cells to lyse. Sadly, the Wollmans were deported from France and perished in Auschwitz. After the end of World War II, their former colleague Andre Lwoff continued their work. By 1949 he had shown that some bacteriophage could exist stably within bacteria in a nonlytic (lysogenic) state. This latent form of the virus (prophage) could be inherited during bacterial division and later be re-activated to produce virus in the absence of any additional infection. Careful studies eventually showed that lysogenic bacteriophage DNA becomes integrated into the genomic DNA of infected bacteria. Much of this latter work was done by François Jacob and by Elie Wollman, the surviving son of Eugene and Elizabeth.
Temin was reportedly unaware of this work because Delbrück (with whom Elie Wollman had trained) and his colleagues at Caltech did not believe in lysogeny. However, by 1962 Vogt and Dulbecco had provided evidence via nucleic acid hybridization that polyoma, a lytic DNA tumor virus originally discovered in mice, was retained in a nonlytic form in oncogenically transformed hamster cells. Soon thereafter, Temin presented similar evidence for the incorporation of the RSV genome into infected cells, although others questioned his results. Temin's proposal that RSV becomes integrated into the DNA of an infected chicken cell genome as a provirus was met with intense skepticism for another reason.
The genome of RSV was known to be composed of RNA, not DNA. Studies of other viruses like polio and influenza had provided examples of RNA serving as a template for the production of more RNA. However, it was generally believed that the flow of information from DNA to RNA to protein was unidirectional. Temin's provirus hypothesis required the heretical conversion of viral RNA into DNA prior to integration and stable inheritance as part of host cell genomic DNA. Temin used inhibitors of DNA synthesis, RNA transcription from DNA, and protein synthesis to obtain evidence consistent with his hypothesis. Viral infection required DNA synthesis, but not protein synthesis. By contrast, viral production by infected cells required RNA synthesis but not DNA synthesis ( Fig. 7 ). Further evidence for a DNA intermediate in viral replication was provided in 1970 by the experiments of David Boettiger in the Temin laboratory. He showed that incorporation of 5-bromodeoxyuridine, a thymidine analog, into the virus caused it to become sensitive to inactivation by ultraviolet light. Similar experiments were also reported by Piero Balduzzi, John Bader, and Herbert Morgan.

Experimental evidence for Temin's provirus hypothesis. Viral infection was prevented by inhibitors of DNA synthesis, but not inhibitors of protein synthesis. By contrast, virus production was prevented by an inhibitor of RNA transcription (actinomycin D), but not by inhibitors of DNA synthesis. (Reprinted from Temin H. 1972. Proc Natl Acad Sci 69: 1016–1020.)
Meanwhile, the criticism had been fierce and unrelenting. Michael Bishop, who would eventually propose his own controversial hypothesis about RSV, described his first encounter with Howard Temin at a scientific meeting in 1968 as follows:
The hypothesis had earned him little but ridicule and grief. So that summer evening, I watched with interest (and from a respectful distance) as Howard argued long into the night with skeptics and detractors. It was my first experience with a scientist who was essentially alone in his beliefs. What I witnessed was a lesson for a lifetime. The opposition to the provirus hypothesis that evening was strong, even vitriolic. In response, Howard was unfailingly patient and reasoned. He had no doubt that his hypothesis was correct, but he was open to constructive criticism, and he painstakingly tried to refute each opposing argument, even those that had no force other than their animus.
Eventually Temin proved to be right. In 1970 he and Satoshi Mizutani reported the existence of an enzyme within detergent-disrupted RSV virions that could indeed convert RNA into DNA. An RNA-dependent DNA polymerase was independently discovered in a murine leukemia virus by David Baltimore, a virologist who had also trained with Renato Dulbecco and was studying the mechanisms of replication of different types of animal viruses. The result was a scientific earthquake, similar in magnitude to that of New Madrid in 1812, which was said to have caused the Mississippi River to run backward. Because of the unprecedented reversal of flow of genetic information from RNA to DNA, this new viral enzyme became known as “reverse transcriptase.” The RNA tumor viruses that encode this enzyme became known as “retroviruses.”
Physical proof of the existence of a DNA provirus came shortly thereafter. Jan Svoboda, a talented virologist, had persisted in studying the biology of RSV behind the Iron Curtain in what was then Czechoslovakia. In the early 1960s he and his colleagues had been able to transform rat fibroblasts by coculture with RSV-infected chicken cells. However, the rat cells themselves were unable to produce infectious RSV unless fused to normal chicken cells. Although these findings supported the provirus theory, Temin himself remained skeptical of this evidence. In 1972, using the same methods and logic by which Oswald Avery and colleagues had first shown that DNA was the genetic material of bacteria, Miroslav Hill and Jana Hillova sealed the deal. They showed that purified genomic DNA from rat cells transformed by RSV could be introduced into uninfected chicken cells, resulting in the production of infectious RSV.
In 1975, Temin and Baltimore received a Nobel Prize in Physiology or Medicine for the discovery of reverse transcriptase, a prize they shared with their mentor Renato Dulbecco for his (and Marguerite Vogt's) work on polyomavirus. Along with the awarding of a Nobel Prize to Peyton Rous in 1966 for the discovery of RSV, this event prompted Peter Duesburg (a fellow retrovirologist) to quip, “One sick chicken, two Nobel prizes.” But that was hardly the end of the story.
From the forthcoming volume Stalking the Enemy Within: A History of Cancer Research , by Joseph Lipsick
Additional Perspectives on A History of Cancer Research available at www.cshperspectives.org
SUGGESTED READING
The phage school, cell culture, and the birth of quantitative animal virology.
- Dulbecco R, Vogt M. 1954. Plaque formation and isolation of pure lines with poliomyelitis viruses . J Exp Med 99 : 167–182. 10.1084/jem.99.2.167 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Eagle H. 1955. Nutrition needs of mammalian cells in tissue culture . Science 122 : 501–514. 10.1126/science.122.3168.501 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Judson HF. 1996. The eighth day of creation: makers of the revolution in biology . Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [ Google Scholar ]
- Skloot R. 2010. The immortal life of Henrietta Lacks . Crown/ Random House, New York. [ Google Scholar ]
- Watson JD, Stent GS, Cairns J. 2000. Phage and the origins of molecular biology . Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [ Google Scholar ]
The Discovery and Assay of RNA Tumor Viruses
- Coffin JM, Hughes SH, Varmus HE editors. 1997. Retroviruses . Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [ PubMed ] [ Google Scholar ]
- Ellermann V, Bang O. 1909. Experimental leukemia in chickens II . Z Hygiene Infektionskrakheiten 63 : 231–272. [English translation available in Some classics in experimental oncology (ed. Shimkin M), NIH Publication No 80-2120 (1980)]. 10.1007/BF02227892 [ CrossRef ] [ Google Scholar ]
- Rous P. 1910. A transmissible avian neoplasm (sarcoma of the common fowl) . J Exp Med 12 : 696–705. 10.1084/jem.12.5.696 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Rous P. 1911. Transmission of a malignant growth by means of a cell-free filtrate . J Am Med Assoc 56 : 198. [ Google Scholar ]
- Temin HM, Rubin H. 1958. Characteristics of an assay for Rous sarcoma virus and Rous sarcoma cells in tissue culture . Virology 6 : 669–688. 10.1016/0042-6822(58)90114-4 [ PubMed ] [ CrossRef ] [ Google Scholar ]
The Discovery of Reverse Transcriptase
- Baltimore D. 1970. RNA-dependent DNA polymerase in virions of RNA tumour viruses . Nature 226 : 1209–1211. 10.1038/2261209a0 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Temin HM, Mizutani S. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus . Nature 226 : 1211–1213. 10.1038/2261211a0 [ PubMed ] [ CrossRef ] [ Google Scholar ]
The Provirus Hypothesis
- Baltimore D. 1995. Thinking about Howard Temin . Genes Dev 9 : 1303–1307. 10.1101/gad.9.11.1303 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cooper GM, Temin RG, Sugden B editors. 1995. The DNA provirus: Howard Temin's scientific legacy . ASM Press, Washington, DC. [ Google Scholar ]
- Hill M, Hillova J. 1972. Virus recovery in chicken cells tested with rous sarcoma cell DNA . Nat New Biol 237 : 35–39. 10.1038/newbio237035a0 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Svoboda J. 2008. The turns of life and science . Adv Cancer Research 99 : 1–32. 10.1016/S0065-230X(07)99001-6 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Temin HM. 1976. The DNA provirus hypothesis . Science 192 : 1075–1080. 10.1126/science.58444 [ PubMed ] [ CrossRef ] [ Google Scholar ]
The Search for Human Tumor Viruses
- Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt's lymphoma . Lancet 283 : 702–703. 10.1016/S0140-6736(64)91524-7 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Javier RT, Butel JS. 2008. The history of tumor virology . Cancer Res 68 : 7693–7706. 10.1158/0008-5472.CAN-08-3301 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Moore PS, Chang Y. 2010. Why do viruses cause cancer? Highlights of the first century of human tumour virology . Nat Rev Cancer 10 : 878–889. 10.1038/nrc2961 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wade N. 1971. Special virus cancer program: travails of a biological moonshot . Science 174 : 1306–1311. 10.1126/science.174.4016.1306 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zur Hausen H. 2009. The search for infectious causes of human cancer: where and why? Virology 392 : 1–10. 10.1016/j.virol.2009.06.001 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Election Results
- The Political Beat
- Latest Links
- Continuing Coverage
- Talking About Race
- Power Grid Security
- Black History in the Carolinas
- Streaming/Mobile Apps
- Newsletter Sign-up (Opens in new window)
- Severe Weather Resources
- Interactive Radar
- 7-Day Forecast
- Tracking the Tropics
- Hour by Hour
- Video Forecast
- School Closings
- Report Closings
- Closings Instructions
- WSOCTV Weather Apps
- Live Stream
- WSOC 24/7 News
- Weather 24/7
- NeighborhoodTV
- The $pend $mart Stream
- Law & Crime
- Curiosity NOW
- 9 Investigates
- Carolina Panthers
- Charlotte FC
- High School Football
- Black History Month
- Carolinas Get Real
- Priced Out Of Charlotte
- Mental Health Resources
- Faces of Pride
- Family Focus
- 9 Food Drive
- 9 School Tools
- Steves Coats
- 9 Crisis Help
- Carolina Strong
- COVID-19 Community Resources
- Back to School
- Steals and Deals
- Contests (Opens in new window)
- Advertise with Us
- Daily 2 Video
- Toyota of North Charlotte
- What's on Channel 9
- What's On TV64
- Closed Captioning
- Visitor Agreement
- Privacy Policy
- Celebrando la Herencia Hispana
- Programas de Telemundo
- Share Your Pics!
Carolina Strong: Woman makes strides in breast cancer community in honor of late mother
CHARLOTTE — Laura Renegar’s mother was just 61 when she lost her third battle with breast cancer.
More than a decade later, Renegar found herself on a life-changing path by walking in the American Cancer Society’s Making Strides Against Breast Cancer event .
She said it was something she always wanted to do in honor of her mother.
“She was amazing. She was the best grandma in the entire world,” Renegar said. “And we did the walk in 2008, 2009, and 2010. And then I was diagnosed.”
By the time of her diagnosis, Renegar said she had become a leader on her team.
“People looked at me and thought, ‘That team leader, she could be diagnosed; I could be diagnosed.’ And when I started hearing that, I decided to take that and push forward,” Renegar said.
As a survivor, she said she has become a force in the breast cancer community.
“We started off our first year; I think we raised $10,000. And we had 50 walkers, and our highest team membership has been 375 walkers. And we’ve raised a million and a half dollars so far,” she explained.
>> Renegar explains the various ways she has helped hundreds of newly diagnosed women, in the video at the top of the page.
Every day, there are people across the Carolinas doing extraordinary things. They’re giving back, they’re helping each other, and they’re making a real difference. We’re highlighting the best in our community in our series, Carolina Strong.
Know somebody who’s making a positive impact? Let us know here .
Carolina Strong: Student raises money for cancer research in honor of mother’s leukemia diagnosis
©2024 Cox Media Group

This job can pay $100K with no degree, and 1,800 spots are available. Hiring opens for 4 days.
:quality(70)/cloudfront-us-east-1.images.arcpublishing.com/cmg/VNL2JUA24RGODGKCDCZCYDOUEM.png)
NC A&T student from Gastonia killed in hit & run, family says
:quality(70)/cloudfront-us-east-1.images.arcpublishing.com/cmg/LNKSVG7XZZDCBLVZWQF2NQUX2Y.png)
Deadly wreck shuts down I-85S in Gaston County
:quality(70)/cloudfront-us-east-1.images.arcpublishing.com/cmg/C6S3J3GOC5H3JGOBOWALACO4AA.png)
Man reported missing after going on fishing trip
:quality(70)/cloudfront-us-east-1.images.arcpublishing.com/cmg/FXATTRGY45CMFLPLIY3M3ABUR4.jpeg)
UNC Charlotte to gain higher status
Health for all, Hunger for none
- English (EN)

Bayer is a global enterprise with core competencies in the Life Science fields of health care and agriculture.
- Our Mission & Strategy
- Our Contributions
- Number of Women in Management Positions
- Overview and Tasks
- Bill Anderson
- Wolfgang Nickl
- Stefan Oelrich
- Heike Prinz
- Rodrigo Santos
- Heiko Schipper
- Julio Triana
- Offices Held at other Companies
- Compensation
- Division Leadership
- Working At Bayer
- Bayer Worldwide
- 2023 - Present
- Biographies
- The Bayer Cross
- The Corporate Archives
- Our Approach
- Supplier Code of Conduct
- Strategic Sustainability Focus Areas
- Supplier Diversity
- Industry Initiatives
- Conditions of Purchase
- Customs & International Trade Guidelines
- Downloads & Links
- Contact Procurement
- Information & IT Security
- Data Privacy Information For Specific Processing Activities
- Compliance Hotline

As a leader in healthcare, Bayer provides innovative solutions designed to prevent, alleviate and treat diseases.
- Health at Bayer
- Heart Attack
- Diagnosing Heart Disease
- Heart Failure
- High Blood Pressure
- Stroke & Atrial Fibrillation
- Gastrointestinal Stromal Tumor
- Breast Cancer
- Kidney Cancer
- Liver Cancer
- Prostate Cancer
- TRK Fusion Cancer
- Thyroid Cancer
- Chronic Kidney Disease
- Age-related Macular Degeneration
- Diabetic Macular Edema
- Healthy Aging
- Contraception
- Endometriosis
- Heavy Menstrual Bleeding
- Uterine Fibroids
- Acne Therapy
- AAV Therapeutics
- Stem cells - The new age of medical research
- Pharmacogenetics
- Precision Medicine
- United against Parkinson’s Disease
- Unlocking the Undruggable Space
- Worldwide Standards
- Ethics in Clinical Trials
- Access to investigational medicines
- Artificial Intelligence
- Chemoproteomics
- PROTAC's
- RNA-targeting SMOLs
- Diagnostic Imaging
- Chemical Probes For Open Access
- Innovation powered by people
- Covalent binding – Vividion
- Immuno-oncology
- Targeted Alpha Therapy
- Tumor Intrinsic Pathways
- Gene Therapy
- Cell Therapy
- Gene Editing
- Patient Access Charter
- Leadership Perspective
- Strengthening Healthcare Access
- Boosting Family Planning Usage through Digital Channels
- Capacity building: Addressing Root Causes through Partnerships
- Impact at Scale: The Challenge Initiative
- Promoting Awareness: World Contraception Day (WCD) & the Your Life Campaign
- Providing Accessible and Affordable Contraceptives
- Enabling Family Planning in Humanitarian Settings
- Chagas Disease
- Moving Non-Communicable Diseases Care Forward
- Ensuring a Sustainable Product Supply
- Delivering Better Cancer Care
- Patient Organization
- Patient Engagement
- News & Stories
- Clinical Trials Explorer
- Transparency Policy
- Aiding Digestion
- Caring for Skin
- Fighting Pain
- Protecting Heart Health
- Relieving Allergies
- Supporting Nutrition
- Treating Cold and Flu Symptoms
- Partnerships
- Trust and Credibility
- Science-Led Self-Care
- Creative and Responsible Marketing
- Empowering Personal Health Sustainably
- Inspiring Creativity and Improving Health
- The Nutrient Gap Initiative
- Report a Side Effect
- EU Verification Logo against Counterfeit Drugs
- Background Information
- Recognizing Fraudulent Brands
- New Safety Features for Prescription Medicine in Europe
- Commitment to Fighting Counterfeit Drugs

Bayer provides tailored solutions for farmers to plant, grow and protect their harvests using less land, water and energy.
- Agriculture Biologicals
- Weed Management
- Disease Management
- Insect Management
- Biotechnology
- Genome Editing
- Plant Breeding
- Smart Solutions
- Bayer and Microsoft Partnership
- Fruits & Vegetables
- Recognize & Avoid Counterfeits
- Bayer´s Role In Combating Counterfeits
- Bayer Safety Seal & Bayer Seal Scan App
- Product Responsibility
- Safe Use Ambassador
- Product Supply
- Farms of the Future
- Innovation Showcase
- Advances in Agriculture
- Innovation Pipeline
- Global LifeHub Network
- Regenerative Agriculture
- Sustainability Commitments
- Reporting Matters
- Our Carbon Zero Commitment
- U.S. Carbon Program
- European Carbon Program
- PRO Carbono Brasil
- GHG Progress Reporting
- Environmental Impact Reduction Reporting Results
- Restoring Biodiversity
- Soil Health
- Land Preservation
- Impact on Smallholder Livelihoods
- Smallholder-Centric Solutions
- Collaboration in Value-Chain Partnerships
- Food Chain Partnership
- Food Loss & Waste
- Food Security
- Food Safety
- Genetically Modified Crops and Bayer
- Farmer Voice
- Crop Protection
- Bayer Innovation
- Youth Ag Summit

Innovative products for the health of humans and plants.
- Product Areas
- Information for Patients
- Beware of Counterfeits
- Report a Side-Effect
- Products from A to Z

For more than 160 years, Bayer has been driving innovation to improve people's lives.
- Conserving Natural Resources in Agriculture
- Rewriting the Future of Healthcare
- Leaps by Bayer
- Research and Development

Read more about our economic, ecological and social challenges and opportunities.
- Latest Updates
- Water Campaign
- Sustainability Report
- Reports and Regulations
- ESG Ratings and Rankings
- Climate Protection
- Water Stewardship
- Environmental Protection
- Health and Safety
- Human Rights
- Bayer Sustainability Council
- Bayer Bioethics Council
- Stakeholder Dialogue
- UN Global Compact
- Sustainability Policy
- Corporate Compliance Policy
- Bayer Human Rights Policy
- Responsible Marketing & Sales
- Bioethical Principles
- Protection of Biodiversity
- Modern Slavery Statement
- Position on Global Product Strategy
- Position on Responsible Care
- Position on Deforestation and Forest Degradation
- Position on Insect Decline
- Raising the Bar on Crop Protection Safety Standards
- UN Sustainable Development Goals
- Position on Sustainable Beef Production
- Bayer Water Position
- Commitment & Overview
- Political Engagement
- Code of Conduct for Responsible Lobbying
- Collaboration with Healthcare Professionals
- Our Disclosure Obligations
- Science Collaborations
- Pharmaceuticals & Medical Devices
- Crop Protection Products & Seeds
- Crop Science ESG Investor Webinar October 2023
- ESG Investor Update Webinar March 2023
- UN Global Compact Adherence Webinar June 2022
- ESG Investor Engagement Activities
- Transparency Reporting
- Science Education
- Culture of Remembrance
- Bayer Clubs
- Bayer and Sports
- Successful Bayer Athletes
- Promotion of New Talent
- Bayer 04 Leverkusen

Besides news this area provides further useful information like interesting stories, visual assets, an event calendar and media contacts.
- Visual Assets
- Social Media
- Media Contacts

Investing in a stronger future - for our shareholders, and for the world.
- Events Archive
- Notice of the Meeting & Agenda
- Stockholders’ Portal
- Countermotions and Proposals for Election
- Stockholder Statements
- Right to speak
- Right of information
- Right to object
- How to Vote
- ASM-Services & -Downloads
- ASM Archive
- Quarterly Reports
- Annual Reports
- Bayer AG Financial Statements
- Compensation Reports and Compensation Systems
- Stock Chart
- Analyst Consensus
- Return Calculator
- ADR Program
- ESG Ratings
- Duties and Activities of the Board of Management
- Supervisory Board
- Disclosure of Managers' Transactions
- Approach to Tax
- Corporate Compliance
- Links & Downloads
- Document Download Tool

Looking for a job in an innovative company? Learn more about Bayer and the opportunities available.
- Our culture
- Your benefits
- Your career
- Follow your dreams
- I belong here
- How to join us
Everything you need to know about Glyphosate and RoundUp safety

- EPA's Review of Glyphosate Safety
There is much discussion about glyphosate, the active ingredient in most Roundup® brand herbicides and other weed-control products. Is it safe to use? What impact does it have on the environment? And more.
By sharing fact-based information and scientific findings from numerous independent sources, we want to show you that glyphosate is safe to use as directed and does not cause cancer in humans.
Glyphosate-based herbicides are among the most widely-used crop protection products. For 50 years, it has been approved and used safely in modern agriculture and food production, vegetation management, lawn care, gardening and more.
Due to its extensive history, rigorous testing and oversight, it’s one of the most studied herbicides in the world.
All crop protection products, including glyphosate, are subject to rigorous testing and oversight by regulatory agencies worldwide. Regarding safety assessments, glyphosate-based herbicides are among the most extensively tested products, with more than 1,500 studies and 50 years of research. After reviewing the volume of scientific research and evaluations by regulatory agencies over the years, experts and regulators worldwide have concluded that glyphosate-based products can be used safely as directed.
As the population grows, the agricultural industry is continuously working to grow healthy crops with less impact on the environment. That means using less land and natural resources, preserving biodiversity, reducing greenhouse gas emissions and helping to ensure that soil stays rich with nutrients.
Providing farmers with tailored solutions designed to address these challenges is crucial. Glyphosate-based products provide efficient and safe crop protection.
Bayer stands fully behind our glyphosate-based Roundup® products, which have been used safely and successfully around the world for 50 years.
Glyphosate-based herbicides are the most widely used and extensively tested herbicides on the market, which is a major reason why so many farmers and others around the world continue to rely on these products not only for effective weed control, but also to minimize tillage farming practices, reduce greenhouse gas emissions, preserve more land for native habitats, and provide enough food to meet the needs of a growing population worldwide.
Like all herbicides, glyphosate has been subject to rigorous testing and oversight by regulatory authorities. The leading health regulators around the world have repeatedly concluded that glyphosate-based products can be used safely as directed. Most recently, in November 2023, the EU Commission re-approved glyphosate for 10 years, following the favorable scientific assessments by its health and safety agencies, including the European Chemicals Agency (ECHA) and European Food Safety Authority (EFSA) , which “did not identify any critical areas of concern.” The U.S. Environmental Protection Agency (EPA) and the regulatory authorities in Japan , Australia , Korea , Canada , New Zealand , and elsewhere have also recently reaffirmed that glyphosate-based products can be used safely as directed.
For details about the U.S. litigation: www.Bayer.com/5pointplan .

IMAGES
COMMENTS
Milestones in Cancer Research and Discovery. During the past 250 years, we have witnessed many landmark discoveries in our efforts to make progress against cancer, an affliction known to humanity for thousands of years. This timeline shows a few key milestones in the history of cancer research. 1775: Chimney Soot & Squamous Cell Carcinoma.
The FDA approved BCG (Bacillus Calmette-Guerin), a bacterial vaccine, for the treatment of bladder cancer, based on research conducted by immunologist Lloyd Old, urologic surgeon Harry Herr and immunologist Herbert Oettgen. First investigated in the 1950s by MSK researchers, BCG remains a primary treatment for non-muscle invasive bladder cancer.
The history of cancer describes the development of the field of oncology and its role in the history of medicine. Early diagnosis ... In 1973, cancer research led to a cold war incident, where co-operative samples of reported oncoviruses were discovered to be contaminated by HeLa.
The History of Cancer. The study of cancer, called oncology, is the work of countless doctors and scientists around the world whose discoveries in anatomy, physiology, chemistry, epidemiology, and other related fields made oncology what it is today. Technological advances and the ever-increasing understanding of cancer make this field one of ...
Learn about the history of cancer, from its discovery in ancient Egypt to the developments that lead to modern treatment. ... The 20th century was an exciting time in cancer research. Carcinogens, chemotherapy, radiation therapy, and better ways to diagnose cancer were all discovered in these years. Some of the most important discoveries of the ...
History of Cancer Research. Cancer research is a vast field of medicine, with the oldest reference found to be of Le Clerc who suggested in 1727, to cut out swellings, polyps, and tumefactions before they became cancerous. Giovanni Morgagni of Padua pioneered the technique of doing autopsy to link the patient's disease to postmortem ...
As part of our dedication to preserving the record of cancer research, and to celebrate our 100th Anniversary, the AACR has commissioned a series of historical review articles to be published serially in our flagship journal, Cancer Research. This history series is intended to cover the major scientific advances of the past 100 years in the ...
The International Journal of Cancer is a cancer journal from the Union for International Cancer Control covering experimental and clinical cancer research. This mini-review chronicles the history of cancer ranging from cancerous growths discovered in dinosaur fossils, suggestions of cancer in Ancient Egyptian papyri written in 1500-1600 BC ...
Cancer research until the 1980s was dominated by a tumour-centric view 7 postulating that mutations in oncogenes and tumour suppressor genes were sufficient to determine carcinogenesis and cancer ...
When Cancer Research UK material is used for commercial reasons, we encourage a donation to our life-saving research. Figure 3 Patient has given verbal consent to use anonymised and slightly modified case history and images for teaching and academic purposes.
1907-1915: The Founding Years. [ Timeline of events, 1907-1915] The American Association for Cancer Research was founded in 1907 at a propitious time. European scientific advances in the late 19th century had set the stage for progress against cancer, a disease that had long been considered hopeless.
Abstract. Observations of the incidence of tumors among chimney sweeps in the eighteenth century and later experiments with coal tars provided early evidence that carcinogens in the environment can promote cancer. Subsequent studies of individuals exposed to radiation, work on fly genetics, and the discovery that DNA was the genetic material ...
So it's no surprise that from the dawn of history people have written about cancer. Some of the earliest evidence of cancer is found among fossilized bone tumors, human mummies in ancient Egypt, and ancient manuscripts. Growths suggestive of the bone cancer called osteosarcoma2have been seen in mummies. Bony skull destruction as seen in ...
Here, we leverage the PCAWG dataset to characterize the evolutionary history of 2,778 cancer samples from 2,658 unique donors across 38 cancer types. We infer timing and patterns of chromosomal ...
Fifteen years ago, the American Association for Cancer Research (AACR) marked its 100th anniversary with the launch of Landmarks in Cancer Research 1907 - 2007, a historical timeline of the seminal scientific discoveries and events that occurred throughout the AACR's first century of existence. The ensuing fifteen years have brought a rapid ...
The legislation raised the federal minimum age for sale of tobacco products, including e-cigarettes, from 18 to 21. (329) Number of cancer survivors in the U.S. reaches 16.9 million. (330,331) AACR membership passes 45,000. AACR Women in Cancer Research (WICR), a membership group within AACR, celebrates 20 years.
History of cancer research has always been closely linked with the history of biology. The rise of the cell theory in the middle of the nineteenth century led to a unified conception of cancer, and to a dramatic change in the description of this disease, as well as in its diagnosis and prognosis. Cancer research has been revitalised during the ...
Abstract. Tumor suppressor genes encode critical intracellular regulators, such as the retinoblastoma protein. They control processes including cell proliferation, cell survival, and responses to DNA damage and are frequently mutated in cancer. In this excerpt from his forthcoming book on the history of cancer research, Joe Lipsick looks back ...
Her husband's cancer and their search for cancer information online inspired CancerHelp UK. History. To begin with, the Medical School server at the University of Birmingham hosted CancerHelp UK. The management of the site passed to The Cancer Research Campaign in 2000. In 2002, The Cancer Research Campaign and the Imperial Cancer Research Fund ...
1916. AACR begins publishing The Journal of Cancer Research, the first English language cancer journal. Oophorectomy decreases breast cancer in mice. Removal of the ovaries from female mice of a strain with a high incidence of spontaneous breast cancer resulted in a decrease in tumors. Later work published in The Journal of Cancer Research ...
Summary:. Research on precancers, as defined as at-risk tissues and early lesions, is of high significance given the effectiveness of early intervention. We discuss the need for risk stratification to prevent overtreatment, an emphasis on the role of genetic and epigenetic aging when considering risk, and the importance of integrating macroenvironmental risk factors with molecules and cells in ...
Diana Azzam is an Assistant Professor and Research Director of the newly established Center for Advancing Personalized Cancer Treatments (CAPCT) at Florida International University.
In this short episode, we delve into Fox's remarkable journey—a marathon across Canada to raise funds for cancer research, despite battling cancer himself.Discover the indomitable spirit of Terry Fox as he ran over 5,000 kilometers on his prosthetic leg, capturing the hearts of Canadians and people worldwide.
A History of Cancer Research: Tumor Viruses. Early studies of transmissible tumors in chickens provided evidence that viruses such as avian leukosis virus (ALV) and Rous sarcoma virus (RSV) can cause cancer in these animals. Doubts about the relevance to human tumors and failures to replicate some early work meant the field of tumor virology ...
As a survivor, she said she has become a force in the breast cancer community. "We started off our first year; I think we raised $10,000. And we had 50 walkers, and our highest team membership ...
Due to its extensive history, rigorous testing and oversight, it's one of the most studied herbicides in the world. ... European Food Safety Authority, International Agency for Research on Cancer (IARC), World Health Organization, Canadian Pest Management Regulatory Agency, Australian Pesticide and Veterinary Medicines Authority, European ...