- Research article
- Open access
- Published: 19 April 2020

Antibiotic use and resistance: an unprecedented assessment of university students’ knowledge, attitude and practices (KAP) in Lebanon
- Samer Sakr 1 ,
- Ali Ghaddar 2 ,
- Bassam Hamam 1 &
- Imtithal Sheet 1
BMC Public Health volume 20 , Article number: 535 ( 2020 ) Cite this article
10k Accesses
20 Citations
Metrics details
The emergence and spread of pathogenic bacteria that is resistant to antibiotics has become a major public health concern. The incorrect prescription, inappropriate consumption and excess use of antimicrobial drugs, specifically antibiotics, are possibly the main factors contributing to the widespread of antibiotic resistant bacteria. This study aims to evaluate the knowledge, attitude and practices (KAP) towards the use of antibiotics as well as their resistance among Lebanese university students in health and non-health related majors.
This cross-sectional study was conducted between May and June 2019 in Beirut (Lebanon) in which 750 students completed a questionnaire made up of four dimensions: Socio-demographic characteristics, 3 questions; assessment of knowledge, attitude and practices, 7, 10 and 1 question, respectively. The data was collected in spreadsheets and analysed with descriptive statistics. The difference in mean scores in each of the knowledge, attitude and practices dimensions between health and non-health related major students was analysed using t-student tests and the difference in percentages using chi-square tests.
Almost 78% of respondents from the health related majors scored high knowledge compared to only 41% of non-health related majors (mean = 4.26; standard error = 0.05 versus mean = 3.41; standard error = 0.13, respectively). The attitude score of the health related major students (35.42%) was positive and more satisfactory compared to the non-health related students (7.32%); (mean = 9.34; standard error = 0.05 versus mean = 9.10; standard error = 0.21, respectively). However, the difference in the scores of attitudes was not statistically significant.
Conclusions
Interventions to promote awareness in this area should focus more students in on non-health related majors.
Peer Review reports
Over the last 5 decades, antibiotics have proven to be an effective and decisive weapon against several diseases. Today, the emergence of pathogenic bacteria that have become resistant to antibiotics, and their spread in the human population, is a growing problem worldwide presenting a significant threat to public health in the twenty-first century, particularly in the developing countries [ 1 , 2 ]. Self-medication, incorrect prescription, inappropriate consumption and excessive use of these antimicrobial drugs could be the key factors for the increase and spread of antimicrobial resistance (AMR) in addition to other equally important social and cultural factors [ 2 , 3 , 4 , 5 ]. This increase in antibiotic resistance will eventually diminish their therapeutic effectiveness and increase treatment failures leading to more severe illnesses with higher mortality rates [ 6 ]. Not to mention the heavy burden this will have on the global economies as well as the different healthcare management systems [ 7 ].
Antibiotic self-medication has become a serious concern and a leading cause of antibiotic resistance. Antibiotic self-medication can result from many factors, such as poor public knowledge and attitude towards antibiotics, easy access to antibiotics in many places and lack of awareness policies on appropriate antibiotic usage [ 8 ]. Numerous studies have reported improper antibiotic use among university students due to self-medication and lack of adequate knowledge of antibacterial agents. Specifically, their indications, their specificity to pathogens and the compliance to dosage regiments [ 9 ].
The assessment of knowledge, attitude and practices (KAP) on a representative sample of university students could be an efficient tool to help improve the use of antibiotics [ 10 ]. Jairoun and colleagues (2019), after conducting a KAP study related to the use of antibiotics among university students in the United Arab Emirates, recommended in their conclusions the development of comprehensive programs and effective educational interventions to remediate the gap in the medical curriculum leading to self-medication practice [ 8 ]. In the same year, Al-Salih and colleagues, after conducting a study about knowledge and attitude regarding antibiotic use among nursing and dentistry students in Babylon University (Iraq), have concluded that despite their knowledge about the appropriate antibiotic use, students lacked in the appropriate attitude [ 11 ]. The same conclusion has been reached regarding professional Indian medical students by Khajuria and colleagues in 2018 [ 12 ]. A cross-sectional questionnaire based study conducted among 2500 Chinese students regarding their KAP of antibiotics concluded that the medical curriculum improves the students’ knowledge on antibiotics. However, since senior medical students showed excessive use of antibiotics, this indicated a lack of appropriate instructions on antibiotic use in their curriculum [ 13 ]. The attitude toward antibiotic use and resistance was average among students of International university of Africa (Sudan) despite having good knowledge as reported by Sunusi and colleagues in 2019 [ 9 ].
Chamoun et al. have reported in 2016, based on a study about the prevalence of antibiotic resistance in 76,278 isolated bacterial strains obtained from 16 Lebanese hospitals (between the years 2013 and 2016), that antimicrobial resistance is becoming a major problem in Lebanon [ 14 ]. In 2017, a study conducted by El Khoury and collaborators, concluded that the low educational and socioeconomic levels of parents as factors significantly associated with poor knowledge and misuse regarding antibiotics [ 15 ]. Cheaito and colleagues, in 2014, conducted a survey among buyers of antibiotics in pharmacies. Their results show that 42% of the participants reported purchasing antibiotics without a prescription. Whereas, almost 19% of the respondents, declared referring to the advice of the pharmacist. Almost 40% justified these practices as a way to save money [ 16 ].
To our knowledge, the evaluation of KAP about rational use of antibiotics among university students, enrolled into health related majors and non-health related majors, in Lebanon has not been assessed yet. Accordingly, the current study aims to evaluate the knowledge, attitude and practices (KAP) towards the antibiotic use and resistance among the university students with health and non-health related majors.
Study design, procedure and sample size
The current study relies on a cross-sectional questionnaire-based survey conducted among a random sample of students enrolled in the largest private university in Lebanon attended mostly by the middle income population ‘see Additional file 1 for the Questionnaire’. The study population comprises of a total of 1250 students. A simple random sample was taken to include students with all health-related majors (biology, biochemistry, medical laboratory, food science, nutrition and pharmacy) as one group, and non-health-related majors (business, engineering, education, arts and computer sciences) as another group.
Data collection
This cross-sectional survey was conducted during the period between May and June 2019. A structured questionnaire was designed and developed by the research team based on literature review and was adapted to cover all the main key points of the research topic (antibiotic use and resistance). The adopted questions were mainly based on previous studies [ 7 , 8 , 17 , 18 , 19 ] and were slightly adapted to the context of the conditions in Lebanon. A pilot study was conducted among 12 students to assess the reliability and validity of the instrument. Data from the pilot study was excluded from the results, but served for adjusting minor modifications to the questions based on the analysis of the collected comments. The reliability of the questionnaire was assessed by calculating the alpha-Cronbach’s coefficient which were found to be satisfactory for the three dimensions of the questionnaire (knowledge: alpha-Cronbach = 0.68, attitudes: alpha-Cronbach = 0.76 and practices: alpha-Cronbach = 0.71). An electronic link (Google form) of the questionnaire was emailed to approximately 1250 students from different majors who were also encouraged to fill the questionnaire during class sessions by their instructors in order to reduce the information and selection bias. Seven hundred fifty students responded to the questionnaire (response rate = 60%), out of which, 63.60% ( n = 477) were majoring in one of the following health related majors: biology, biochemistry, nutrition, food sciences, biomedical sciences and pharmacy. The remaining 36.4% of students ( n = 273) were in non-health related majors such as: business administration, arts, engineering or education.
The questionnaire included dimensions on the KAP (knowledge, attitude and practices) towards antibiotic use and resistance. The data was collected in excel sheet and analysed with descriptive statistics and results expressed as means and standard deviations, frequencies and percentages. Questions were grouped into four categories reflecting the participants’ socio-demographic characteristics (3 questions including age, gender and education), knowledge (7 questions including, as an example, “Are antibiotics effective to treat urinary tract infection?”), attitude (10 questions including, as an example, Is it okay to buy the same antibiotics, if you are sick and they helped you get better and practices (1 question, “Do you check the expiry date of the antibiotic before using it?”). Participants scored 0 on each question with the wrong answer and 1 for each question with the right answer. The sum of scores was calculated for the two dimensions of knowledge and practices considering the sum of the score for each individual question in each dimension. Scores ranged from 0 to 7 in the knowledge dimension and from 0 to 10 in the attitudes dimension.
Statistical analysis
Descriptive statistics showed the frequency and percentage (%) of participants who answered correctly for the different questions related to socio-demographic characteristics, knowledge, attitude and practices towards antibiotics use. Chi-square test of independence was used to compare frequency of participants who answered correctly between the health and non-health related majors. T-student test was used to compare the average score in the two domains of knowledge and attitude between students in the health and non-health related majors.
Study participants
A total of 1250 participants was randomly selected out of 10,000 students enrolled at the university. Seven hundred fifty students (out of 1250) responded to the questionnaire giving this study a response rate of 60%. Respondents were categorized into two groups based on their respective majors: 63.60% ( n = 477) were majoring one of the following health related topics (biology, biochemistry, nutrition, food sciences, biomedical sciences and pharmacy); while the remaining students 36.4% ( n = 273) were studying a non-health related major (business administration, arts, engineering or education).
Socio-demographic characteristics of participants
As indicated in Table 1 , more than half of the participants 568 (75.73%) were females and 182 (24.26%) were males. The majority of the participants 493(65.73%) were aged 18–21 years; and 161 (21.46%) participants were aged 22–23 years; 95 (12.66%) and one (0.13%) participants were aged more than 23 and less than 18, respectively. The majority of participants 477 (63.60%) were enrolled in health related education and 273 (36.40%) were in non-health related education.
Knowledge of antibiotics use among participants
Table 2 indicates the frequency and % of participants who answered yes/no for each question related to knowledge, attitude and practices towards antibiotics use. The % of students in health-related majors who got the correct answer was higher than those in non-health related majors for the majority of questions related to knowledge (effectiveness of antibiotics for treating viral vs. bacterial infections, for treating urinary tract infections, for treating fever, the right time to stop taking antibiotics and the familiarity with terms related to antibiotic resistance). These differences in scores were statistically significant in most of the questions except for the question related to effectiveness of antibiotics for the treatment of fever. On the other hand, the % of students in non-health related majors who answered correctly was higher in only two questions related to effectiveness of antibiotics to treat malaria and headaches; however, the difference was statistically significant only for the question related to malaria. The biggest difference in the answers of students from the two groups was in the question about effectiveness of antibiotics to treat viral infections, where 80.2% of students in health related majors gave the correct answer compared to only 36.9% of students in the non-health related majors. It is interesting to note that the majority of students in both groups knew when to stop using antibiotics (95 and 80.7% in the health and non-health related majors, respectively). The percentages of participants who got the correct answers in both groups, together with the p -values are also showed in Table 2 .
Attitude towards antibiotics use among participants
The % of participants who gave correct answers in the health related majors was higher than the non-health group on all questions related to attitude except for the question about attitude towards the spread of antibiotic resistance from one person to another where the non-health group scored higher with p -value = 0.07. The difference in scores between the health and the non-health group was statistically significant in all of the questions except for two questions about attitudes towards antibiotic resistance spread due to insufficient knowledge and inappropriate use ( p = 0.21 and 0.17, respectively). The % difference of participants who had the correct attitudes towards antibiotic resistance was the highest in the question about buying the same antibiotics that helped treat the same symptoms in the past (84% vs. 68.2% for health and non-health groups respectively).
Participants were also asked about their attitude whether or not pharmaceutical companies should develop new antibiotics. The % of respondents who believed that pharmaceutical companies should develop new antibiotics was higher in the health-majored students (98.6% vs. 89.6%). A rapid qualitative analysis of the participants’ written responses while answering one question about their subject opinion on the subject matter revealed that both groups (health and non-health) were aware about the importance of developing new antibiotics to deal with issues of resistance. It was noted that some non-health majoring students were aware that “new viruses and infections are born and most cannot be properly treated with existing antibiotics” as given by this example. While, most health related students used more scientific terms to express their opinions as in this example: “Because the bacterial cells will be more resistant to the current antibiotics due to mutations that occur.”
Practices on antibiotics use among participants
When asked about previous use of antibiotics, 50% of all participants answered that they have previously used Augmentin. The second and third most commonly used antibiotics were Flagyl (12%) and Amoxicillin (9%). 10% answered that they have never used antibiotics before.
Practices were assessed through asking participants whether they check the expiry date of the antibiotic before using it. The majority of participants in both groups (84.4% in health and 74.5% in non-health related majors replied that they do check. The % was higher in the health-related major group with statistically significant difference form the non-health group ( p -value≤0.001).
Analysis of overall knowledge and attitude scores
The average knowledge score was higher in the health major group of students compared to the non-health group (mean = 4.26; standard error = 0.05 vs. mean = 3.41; standard error = 0.13, respectively). This difference in scores was statistically significant ( p -value≤0.001). The average attitude score was higher in the health major group of students compared to the non-health group (mean = 9.34; standard error = 0.05 vs. mean = 9.10; standard error = 0.21, respectively). However, the difference in the scores of attitudes was not statistically significant ( p -value = 0.12).
Antibiotic resistance is a serious public health problem. Assessment of knowledge, attitude and practices of antibiotic use in university students can greatly impact how best to tackle the growing threat of the antibiotic resistance and its related issues [ 18 , 20 ]. In the present study, students in health related majors had better knowledge (higher percentages of correct answers) in almost all the questions related to knowledge; and, had more informed attitude towards dealing with the problem of antibiotic resistance (had higher percentages of correct answers in all questions related to attitude). Thus exhibiting a good knowledge, and satisfactory behavioural attitude towards a rational use of antibiotics. The knowledge and attitude of students in non-health majors were, as expected, less satisfactory. Strikingly, both groups of students have shown exemplary attitude when it comes to antibiotic resistance. As for practices, most of the students regardless of their majors were well aware of good practices. Our findings showed that 80.2% ( n = 381) of students in health related majors were aware that antibiotics are used to treat bacterial infections. Whereas 36.9% ( n = 101) of the students in non-health related majors were knowledgeable about the effectiveness of antibiotic against bacterial or viral infection. Moreover, the current study revealed that the large majority of students in health related majors compared to non-health majors (95.2% vs. 80.7, respectively), were knowledgeable about the timing of when to stop the antibiotics use.
In an article published in 2017 by Jamhour et al. including 400 adults’ respondents from two cities in Lebanon, they found that 61% thought that antibiotics should be taken as a common cold treatment. They also showed a significant correlation between self-medication and lower educational level. In addition, the respondents in that study who had lower knowledge about antibiotics, usually stopped antibiotics at the inappropriate time [ 21 ]. Mouhieddine and colleagues have reported in 2015, based on a random convenience sample of 500 people in Lebanon, that 46.1% of them expressed moderate knowledge levels, where 3.5% did not know that antibiotics are not anti-viral. In this study, 56.0% of the respondents also expect the doctor to prescribe an antibiotic for the common cold [ 22 ]. Similarly, Jifar and Ayele in 2018, reported that 83% of respondents in Harar city, Eastern of Ethiopia, replied that antibiotics speed up the recovery colds [ 17 ]. On the other hand, Jairoun and his colleagues have reported in 2019 that the large majority of university students were aware that antibiotics can kill bacteria and can be used to cure bacterial infections [ 8 ]. Moreover, Khajuria et al. 2019 showed that 90% of medical students agreed that antibiotics are useful for bacterial infections [ 12 ]. Gary and colleagues, have reported in 2012, while comparing the KAP related to antibiotic use and resistance among medical and non-medical university students in Jordan, that 44% percent of non-medical students and 28.1% of medical students agreed that antibiotics could cure cold and viral infections [ 23 ]. In Britain, 38% of respondents ignored that antibiotics cannot resolve colds [ 24 ]. Several studies have revealed that antibiotics are more likely to be prescribed under patients’ pressure [ 25 ]. Another study revealed widespread misconceptions about the utility of antibiotics for viral infections [ 26 ]. This is consistent with the findings of a global survey conducted by the world health organization (WHO) in 2015 [ 27 ]. WHO established a key strategy by engaging the prescribers and educating the public to reduce misuse of antibiotic use [ 28 , 29 ].
In our study, the majority of the students enrolled in health related majors (93.3%), were familiar with terms related to antibiotic resistance, whereas around half of non-health related major students (56.6%) were aware of such terms. Jamhour and colleagues published in 2017 that 83% of the 500 respondents in Lebanon knew that the misuse of antibiotics could result in microbial resistance [ 21 ]. In a report published in 2015, it showed that 48.5% of respondents from Lebanon, declared continuing to take their full course of antibiotics even if their symptoms improved, underlying an alarmingly 51.5% who could stop their treatment after symptoms improvement [ 22 ]; similar to what was found by a number of previous studies [ 22 , 30 , 31 ].
In the current study, about half of the university students reported the use of antibiotics at least once in the year prior to study. A study done in Lebanon (2015) showed that 68.3% of the considered sample consumed antibiotics 1–3 times per year [ 22 ]. Our data is more consistent with a study (53.5%) conducted in Harar city, Eastern Ethiopia [ 17 ]. At the same time, our scores were higher than what was reported by Tesfaye (2017) who reported that 35.9% of participants consume antibiotics once during the year preceding the study [ 32 ]. On the other hand, our finding was considerably lower than what was found in Namibia, which was 80% [ 33 ].
The average knowledge score was significantly higher in the health major group compared to the non-health major students ( p -value≤0.001). The average attitudes score was higher in the health major group of students compared to the non-health group. When comparing our data with other studies conducted among university students, our results agree with the survey conducted among medical and non-medical Chinese university students as reported by Huang et al. 2013 where they reported that Medical students were better than non-medical students in terms of attitude, knowledge and perception on the level of public education on antibiotic use, but worse on behavior. However, they found that but senior medical students have more positive behavior on the usage of antibiotics compared with low grade medical students and non-medical students in general [ 13 ]. Same pattern was observed by another study of medical school students scoring remarkably better than those the non-medical school in KAP towards antibiotic use and resistance [ 8 ]. In another study, it was found that 80% of nursing and dentistry students in Babylon University, Iraq have high knowledge but inappropriate attitude [ 11 ], results which were similarly found among Indian and Sudanese medical university students [ 9 , 14 ]. Higuita-Gutiérrez and colleagues, reported in 2020 that medical students from three medical schools in Medellin, Colombia exhibit poor knowledge regarding antibiotic use due to insufficient training with regard to antibiotic use and bacterial resistance [ 34 ]. Whereas, Veses and colleagues in the same year, after surveying undergraduate dental students at Universidad Cardenal Herrera, concluded that awareness campaigns are needed to promote student’s use of antibiotics in young generations particularly among the pre-professional health sciences students [ 35 ]. Interestingly, as a result of the lack of training they discovered, Tsopra et al. 2020 used a game called ‘AntibioGame’ through which students play the role of a doctor meeting patients in consultation as a promising tool for improving knowledge in antibiotic prescription [ 36 ].
Taking all of the above into consideration, as well as our findings about the lack of appropriate knowledge among university students, irrespective of their major, when questioned about the effectiveness of antibiotics to treat urinary tract infection, malaria, and headache or fever (where the correct answers varied between 17.7 and 53.3%), we recommend that awareness programs and educative measures must be better incorporated in students’ curricula to remediate the gaps related to their knowledge about antibiotic use.
As for the attitude assessment, all students, whether they were enrolled in health or non-health related majors, agreed that AMR is a serious public health issue and that repeated use of antibiotics and insufficient knowledge could lead to antibiotic resistance. These findings were similar, though in better numbers, to the previous studies [ 20 , 31 ]. Jifar and Ayele in 2018, published in their study that 78.4% of subjects in Harar city agreed that the unnecessarily use of antibiotics can increase the antibiotic resistance [ 17 ], 69.7% in Ethiopia [ 32 ], 50% in Jordan [ 37 ], and 72% in Namibia [ 33 ]. Our presented results revealed that students with health related major had a favorable and better attitude about rational use of antibiotics compared to the group of students with non-health major. Only 68.6% ( n = 120) of the students with non-health major agreed that “a doctor is a good one even if he does not prescribe antibiotics when the patient thinks that it is needed” whereas, 83.1% ( n = 305) of the opposite group of students share the same opinion. It has been published by Mouhieddine and his colleagues that 65.1% of the 500 respondents questioned in Lebanon in 2015, referred to doctors’ prescriptions regarding antibiotics, and 22.4% declared, alarmingly, that they self-medicate [ 22 ]. In the study of Jifar and Ayele in 2018, most respondent (90%) agreed on the need for physician consultation before purchasing antibiotics and 73.1% declared getting prescription to purchase antibiotics. This finding is just higher than study done in Saudi Arabia in which they reported, 76.6 and 66.6%, respectively for the same questions [ 17 ]. According to Jamhour and colleagues in 2017, it is common that Lebanese get access to antibiotics without a prescription despite the high ratio of physicians to patients in Lebanon [ 38 ]. The same pattern was observed in our study when students of different majors where questioned about the efficiency of using same antibiotics to treat same symptoms faced in a previous disease. Almost 70% of non-health related major compared to 84% of students of the second group answered this question correctly. Surprisingly, significant number of both group of students, 85.2% ( n = 202) and 94% ( n = 435) from non-health and health majoring students respectively, found it unacceptable to use antibiotics from a friend or family member to treat an infection. On the same topic, it has been reported by Jifar and Ayele in 2018 that 87.2% of respondents in Harar city in Ethiopia were aware not to keep antibiotics for future use; 90% also thought that antibiotics should not be shared among friends and family members without prior physician consultation; and 65.3% self-prescribed antibiotics translating poor knowledge and attitude toward antibiotics use [ 17 ]. Another study reported that only 17% of participants kept antibiotics in their home for future use [ 39 ]. A Namibian study reported that 28.5% of users kept antibiotics for future use [ 40 ]. In India, 76% used antibiotics without prescription [ 41 ], 32.7% in Italy [ 42 ], 28.8% in Saudi Arabia [ 43 ], and 9% in Hong Kong [ 44 ]. This difference might be due to variation of regulations and their application from one country to another in addition to differences in the socio-demographical conditions. The findings presented in the current study indicate that students were not well aware of the irrational use of antibiotics, though students in health majors showed a better attitude, this is different from findings published previously [ 45 , 46 ]. Based on our sample, university students were well aware of the development of antibiotic resistance. However, the responses of the students in the present study cannot be generalized to other universities, since students could have different educational programs, skills and experiences [ 47 , 48 ]. It is indeed a striking findings of this study having such a disparity in the attitude towards antibiotic use where 100% of students, regardless their respective majors, are aware of the issue of antibiotic resistance but up to 32%, in some cases, lack the appropriate attitude, particularly among the group of students in non-health related major. In this regards, our findings are similar to those reported by Mouhieddine et al., in 2015 that 40.6% of the 500 respondents demonstrated only moderate attitudes [ 22 ].
Our findings clearly indicate that it is urgent to limit the granted access for antibiotics in Lebanon and other developed countries. Indeed, the WHO is voicing alarms about the increasing levels of the development, worldwide, of antibiotic resistant pathogenic bacteria. In order to remediate to this major issue, the WHO issued a “Global Strategy for Containment of Antimicrobial Resistance” pressing governments and decision makers to apply and take actions as has happened in South Korea where a number of national educational campaigns on the appropriate use of antibiotics in various ways targeting the general public have been implemented [ 49 ]. Our report shows that knowledge and attitude, of university students, towards antibiotic use and antibiotic resistance could be positively impacted, though not always sufficiently, by more specialized course material related to health. This strengthens the need of the inclusion in the curriculum of students in non-health majors of strategies allowing to get familiarized with public health issues.
Our study highlighted the possible need for knowledge-based education programs for students, especially in the non-medical or non-health related fields. Specifically, our suggestions include seminars, workshops and courses in students’ curricula the extent and effectiveness of which can be the aim of future studies. The quick implementation of awareness campaigns about knowledge and appropriate use of antibiotics seems to be a priority based on ours and others findings. In addition, health authorities should expand their investments in policy making and in a more rigorous surveillance system regarding the access to antibiotics. Awareness campaigns could be done in a number of different routes: i) through national strategies promoting vaccination and hygiene; ii) by updating curricula in universities including public health courses/ workshops in all majors; ii) through media campaigns and intervention; and, iv) through a greater proactive role for pharmacists. The absence of such strategies could result in a continuous degradation of the KAP towards antibiotic use and resistance, leading to more serious consequences on the development of AR.
Limitations of the study
The present study has a few limitations. To start with, results should be treated with caution before their generalization to the population of university students in Lebanon or the region since they are based on a cross-sectional design among a random sample of one university in Lebanon. Although the response rate was acceptable (60%), further studies with larger sample sizes and including more university and non-university students are needed to understand better the level of awareness of young adults and adolescents about the issue of antimicrobial resistance in Lebanon and the region.
Antimicrobial resistance is a serious public health problem. Assessment of knowledge, attitude and practices of antibiotic use in university students can greatly impact the antibiotic related issues. This study addressed KAP about use of antibiotics and AR among students enrolled in a large private university in Lebanon. Our findings indicate that improving the students’ level of knowledge about the use of antibiotics might remediate and rationalize their attitude toward antimicrobial use. The curriculum of students with non-health related majors requires improvement to include seminars, workshops, and/or courses related to public health concerns such as AMR. Our recommendations are in line with what has been proposed by several comparable studies [ 7 , 8 , 13 ]. This would increase the knowledge of students with non-health majors towards public health issues. Awareness campaigns through media considering public health is also recommended. Surveillance system restricting the granted accessibility of antibiotics is an urgent need added to the involvement of the clinicians in sharing more efficiently their knowledge with the patients would aid in ensuring rational use of antibiotics and thus control the growing problem of antibiotic resistance. Involving the civil society organisation and the media intervention would greatly serve this aim.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
Antimicrobial resistance
Knowledge, attitude and practices
- Antibiotic resistance
World Health Organization
Organisation Mondiale de la Santé (OMS). Résistance aux antimicrobiens. Aide-mémoire N°194. 2016: http://www.who.int/mediacentre/factsheets/fs194/fr/ . Accessible 13 Février 2017.
Google Scholar
Vadivoo NS, Usha B, Padmavathi BK. Assessment of clinician’s knowledge and perception on antimicrobial resistance a primary strategy for antimicrobial resistance control. Glob J Med Res. 2015;15(4):9–14.
Shehadeh M, Suaifan G, Darwish RM, Zaru L. Knowledge, attitudes and behavior regarding antibiotic use and misuse among adults in the community of Jordan. A pilot study. Saudi Pharm J. 2012;20(2):125–33.
Article PubMed Google Scholar
Center for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States. Atlanta: CDC; 2013.
Sosa A, Byarugaba D, Amabile-Cuevas C, Hsueh P, Kariuki S, Okeke I. Antimicrobial resistance in developing countries. New York: Springer link; 2010.
Book Google Scholar
World Health Organization. WHO Global Strategy for Containment of Antimicrobial Resistance.WHO/CDS/CSR/DRS/2001.2. http://www.who.int/csr/resources/publications/drugresist/en/ EGlobal_Strat.pdf. Accessed 11 July 2014.
Jairoun A, Hassan N, Ali A, Jairoun O, Shahwan M. Knowledge, attitude and practice of antibiotic use among university students: a cross sectional study in UAE. BMC Public Health. 2019;19(1):518.
Article PubMed PubMed Central Google Scholar
Jairoun A, Hassan N, Ali A, Jairoun O, Shahwan M, Hassali M. University students’ knowledge, attitudes, and practice regarding antibiotic use and associated factors: a cross-sectional study in the United Arab Emirates. Int J Gen Med. 2019;12:235–46.
Sunusi LS, Awad MM, Hassan NM, Isa CA. Assessment of knowledge and attitude toward antibiotic use and resistance among students of International University of Africa, medical complex, Sudan. Glob Drugs Therapeutics. 2019;4:1–6.
Pallavi K, Alice K, Regina R, Indla R. An evaluation of knowledge, attitude and practice of rational antibiotic usage and antibiotic resistance among interns in a teaching tertiary care hospital: a cross sectional questionnaire/ based study. Indian J Pharm Pharm. 2017;4(4):192–7.
AL-Salih SSH, Hindi NKK, Abdul Kadhim ZH, Naji ST, Abbas AS, et al. Knowledge and attitudes regarding antibiotic use and resistance among nursing and dentistry students in Babylon University/ Iraq. Indian J Forensic Med Toxicol. 2019;13(4):1147–52.
Article Google Scholar
Khajuria K, Kaur S, Sadiq S, Khajuria V. KAP on antibiotic usage and resistance among second professional medical students. Int J Basic Clin Pharm. 2018;8(1):68–73.
Huang Y, Gu J, Zhang M, Ren Z, Yang W, Chen Y, et al. Knowledge, attitude and practice of antibiotics: a questionnaire study among 2500 Chinese students. BMC Med Educ. 2013;13:163.
Chamoun K, Farah M, Araj G, Daoud Z, Moghnieh R, Salameh P, et al. Surveillance of antimicrobial resistance in Lebanese hospitals: retrospective nationwide compiled data. Int J Infect Dis. 2016;46:64–70.
Article CAS PubMed Google Scholar
El-Khoury G, Ramia E, Salameh P. Misconceptions and malpractices toward antibiotic use in childhood upper respiratory tract infections among a cohort of Lebanese parents. Eval Health Prof. 2017;41(4):1–19.
Cheaito L, Azizi S, Saleh N, Salameh P. Assessment of self-medication in population buying antibiotics in pharmacies: a pilot study from Beirut and its suburbs. Int J Public Health. 2014;59(2):319–27.
Jifar A, Ayele Y. Assessment of knowledge, attitude, and practice toward antibiotic use among Harar City and its surrounding community, eastern Ethiopia. Interdiscip Perspect Infect Dis. 2018. https://doi.org/10.1155/2018/8492740 .
Awad AI, Aboud EA. Knowledge, attitude and practice towards antibiotic use among the public in Kuwait. PLoS One. 2015;10(2):1–15.
Article CAS Google Scholar
Waaseth M, Adan A, Røen IL, Eriksen K, Stanojevic T, Halvorsen KH, Garcia BH, et al. Knowledge of antibiotics and antibiotic resistance among Norwegian pharmacy customers – a cross-sectional study. BMC Public Health. 2019;19:66.
Chandan NG, Nagabushan H. Assessment of knowledge, attitude and practice of interns towards antibiotic resistance and its prescription in a teaching hospital: a cross-sectional study. Int J Basic Clin Pharm. 2016;5(2):442–11.
Jamhour A, El-Kheir A, Salameh P, Abi Hanna P, Mansour H. Antibiotic knowledge and self-medication practices in a developing country: a cross-sectional study. Am J Infect Control. 2017;45(4):384–8.
Mouhieddine TH, Olleik Z, Itani MM, Kawtharani S, Nassar H, Hassoun R, et al. Assessing the Lebanese population for their knowledge, attitudes and practices of antibiotic usage. J Infect Public Health. 2015;8(1):20–31.
Gary S, Shehadeh M, Darwish DA, et al. A cross-sectional study on knowledge, attitude and behavior related to antibiotic use and resistance among medical and non-medical university students in Jordan. Afr J Pharm Pharmacol. 2012;6(10):763–70.
McNulty CA, Boyle P, Nichols T, Clappison P, Davey P. Don’t wear me out- the public’s knowledge of and attitudes to antibiotic use. J Antimicrob Chemother. 2007;59(4):727–38.
Ling Oh A, Hassali M, Al-Haddad M, Syed Sulaiman S, Shafie A, Awaisu A. Public knowledge and attitudes towards antibiotic usage: a cross-sectional study among the general public in the state of Penang, Malaysia. J Infect Dev Countries. 2011;5(5):338–47.
Mo Y, Ivan S, Pei Shi PL, Xiang Lee JK, Kien Yee MW, Kwan Ki KK, Rick Twee-Hee O, Paul AT, Alex RC. Relating knowledge, attitude and practice of antibiotic use to extended spectrum beta-lactamase-producing Enterobacteriaceae carriage: results of a cross-sectional community survey. BMJ Open. 2019. https://doi.org/10.1136/bmjopen-2018-023859 .
World Health Organization. Antibiotic resistance: multi-country public awareness survey. http://www.who.int/drug resistance/documents/baselinesurveynov2015/en/ . Accessed 25 Mar 2018.
McNulty CA, Cookson BD, Lewis MA. Education of healthcare professionals and the public. J Antimicrob Chemother. 2012. https://doi.org/10.1093/jac/dks199 .
Little P, Stuart B, Francis N, et al. Effects of internet-based training on antibiotic prescribing rates for acute respiratory-tract infections: a multinational, cluster, randomised, factorial, controlled trial. Lancet. 2013;382(9899):1175–82.
Khan Afzal AK, Banu G, Reshma KK. Antibiotic resistance and usage- a survey on the knowledge, attitude, perception and practices among medical students of a southern Indian teaching hospital. J Clin Diagn Res. 2013;7(8):1613–6.
CAS Google Scholar
Sampath S, Venoukichenane V. Knowledge, attitude and practice of antibiotic usage among health care personnel in a tertiary care hospital. Scholars J Appl Med Sci. 2016;4(9):3294–8.
Tesfaye Z. Patient knowledge and practice on antimicrobial use and resistance in Felege Hiwot hospital, Bahir Dar, Ethiopia. J Basic Clin Pharm. 2017;8:S010–5.
Pereko DD, Lubbe MS, Essack SY. Public knowledge, attitudes and behaviour towards antibiotic usage in Windhoek, Namibia. South Afr J Infect Dis. 2015;30(4):134–7.
Higuita-Gutiérrez LF, Molina-Garcia V, Acevedo Guiral J, Gómez Cadena L, et al. Knowledge regarding antibiotic use among students of three medical schools in Medellin, Colombia: a cross-sectional study. BMC Med Educ. 2020;20(1):22.
Veses V, Del Mar J-SM, González-Martínez R, Cortell-Ballester I, Sheth CC. Raising awareness about microbial antibiotic resistance in undergraduate dental students: a research-based strategy for teaching non-laboratory elements of a microbiology curriculum. BMC Med Educ. 2020;20(1):47.
Tsopra R, Courtine M, Sedki K, Eap D, Cabal M, et al. AntibioGame®: a serious game for teaching medical students about antibiotic use. Int J Med Inform. 2020;136:104074. https://doi.org/10.1016/j.ijmedinf.2020.104074 .
Darwish DA, Abdelmalek S, Abu Dayyih W, Hamadi S. Awareness of antibiotic use and antimicrobial resistance in the Iraqi community in Jordan. J Infect Dev Countries. 2014;8(5):616–23.
Sovereign and investment reports; Lebanon country profile. http://www.finance.gov.lb/ENUS/FINANCE/REPORTSPUBLICATIONS/DOCUMENTSANDREPORTSISSUEDBYMOF/Pages/SovereignAndInvestmentReports.aspx . Accessed 27 September 2015.
Lim KK, Teh CC. A cross sectional study of public knowledge and attitude towards antibiotics in Putrajaya, Malaysia. Southern Med Rev. 2012;5(2):26–33.
Pavydė E, Veikutis V, Mačiulienė A, Mačiulis V, Petrikonis K, Stankevičius E. Public knowledge, beliefs and behavior on antibiotic use and self-medication in Lithuania. Int J Environ Res Public Health. 2015;12(6):7002–16.
Chandrakanth P, Mohamed Saleem TS, Reddy MM, Gopinath C, Rao MM. Assessment of public knowledge and attitude regarding antibiotic use in a tertiary care hospital. Asian J Pharm Clin Res. 2016;9(1):118–22.
Napolitano F, Izzo MT, di Giuseppe G, Angelillo IF. Public knowledge, attitudes, and experience regarding the use of antibiotics in Italy. PLoS One. 2013;8(12):e84177.
Article PubMed PubMed Central CAS Google Scholar
Abdulaziz Aldhafar S, Talat W. Attitude, and practice toward the usage of antibiotics among public in Al-Ahsa, Saudi Arabia. Int J Sci Stud. 2017;4(11):14–7.
You JHS, Yau B, Choi KC, Chau CTS, Huang QR, Lee SS. Public knowledge, attitudes and behaviour on antibiotic use :a telephone surveying Hong Kong. Infection. 2008;36(2):153–7.
Singh M, Singh AK. Knowledge, attitude and practice study on awareness of antibiotic stewardship among health care professionals in a tertiary care hospital in Delhi. Int J Curr Microbiol App Sci. 2017;6(7):238–45.
Jain A, Dhir KS, Batta M, Singh G. Knowledge and practices in community regarding antibiotic usage. Int J Res Med Sci. 2016;4(2):610–4.
Tevatea S, Chaudhry S, Rath R, et al. A questionnaire based survey on knowledge, attitude and practice of antibiotics among dental and paramedical students – a cross sectional survey. World J Pharm Pharm Sci. 2016;5(5):1205–16.
Desai AJ, Gayathri GV, Mehta DS. Public’s perception, knowledge, attitude and behavior on antibiotic resistance – a survey in Davangere city, India. J Prev Med Holistic Health. 2016;2(1):17–23.
Kim S, Moon S, Kim E. Public knowledge and attitudes regarding antibiotic use in South Korea. J Korean Acad Nurs. 2011;41(6):742–9.
Download references
Acknowledgements
The authors would like to thank all the students at the Lebanese International University (LIU) who have participated in the study.
No funding were used to assist in the preparation of this study.
Author information
Authors and affiliations.
Department of Biological and Chemical Sciences, School of Arts and Sciences, Lebanese International University, Beirut, Lebanon
Samer Sakr, Bassam Hamam & Imtithal Sheet
Department of Biomedical Sciences, School of Arts and Sciences, Lebanese International University, Beirut, Lebanon
Ali Ghaddar
You can also search for this author in PubMed Google Scholar
Contributions
SS and IS designed the study and carried out data collection. AG analyzed and interpreted the data. SS, AG, and BH drafted the manuscript. All authors have approved the final version of manuscript.
Corresponding author
Correspondence to Samer Sakr .
Ethics declarations
Ethics approval and consent to participate.
The study was reviewed and approved by the Lebanese International University Institutional Review Board (IRB) ethical committee (Reference LIUIRB-190424-SS1). Informed written consent to participate was obtained from all the participants included in this study. A consent form was included in the questionnaire, explaining the research objective and assuring that anonymity and confidentiality would be maintained, and that the participation of students is voluntary and would not affect in any way their grades at university.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1..
Questionnaire form.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Sakr, S., Ghaddar, A., Hamam, B. et al. Antibiotic use and resistance: an unprecedented assessment of university students’ knowledge, attitude and practices (KAP) in Lebanon. BMC Public Health 20 , 535 (2020). https://doi.org/10.1186/s12889-020-08676-8
Download citation
Received : 16 October 2019
Accepted : 07 April 2020
Published : 19 April 2020
DOI : https://doi.org/10.1186/s12889-020-08676-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Antibiotic use
- Health related majors
- And non-health related majors
BMC Public Health
ISSN: 1471-2458
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 11 April 2022
Prevalence and characterisation of antimicrobial resistance genes and class 1 and 2 integrons in multiresistant Escherichia coli isolated from poultry production
- Przemysław Racewicz 1 ,
- Michał Majewski 1 ,
- Hanna Biesiada 1 ,
- Sebastian Nowaczewski 1 ,
- Jarosław Wilczyński 4 ,
- Danuta Wystalska 4 ,
- Magdalena Kubiak 3 ,
- Marcin Pszczoła 2 &
- Zofia E. Madeja 2
Scientific Reports volume 12 , Article number: 6062 ( 2022 ) Cite this article
5057 Accesses
19 Citations
Metrics details
- Antimicrobials
- Environmental microbiology
- Microbial genetics
- Public health
A global increase in the populations of drug resistant bacteria exerts negative effects on animal production and human health. Our study has been focused on the assessment of resistance determinants in relation to phenotypic resistance of the 74 commensal E. coli isolates present in different ecological environments. The samples were collected from poultry litter, feces, and neck skin. Among the microorganisms isolated from the poultry litter (group A), the highest resistance was noted against AMP and DOX (100%). In the E. coli extracts from the cloacal swabs (group B), the highest resistance was observed against AMP (100%) and CIP (92%). The meat samples (group C) were characterized by resistance to AMP (100%) and STX (94.7%). Genes encoding resistance to β-lactams ( bla TEM , bla CTX-M ), fluoroquinolones ( qnrA, qnrB, qnrS ), aminoglycosides ( strA-strB, aphA1, aac(3)-II ), sulfonamides ( sul1, sul2, sul3 ), trimethoprim ( dfr1, dfr5, dfr7/17 ) and tetracyclines ( tetA, tetB ) were detected in the studied bacterial isolates. The presence of class 1 and 2 integrons was confirmed in 75% of the MDR E. coli isolates (plasmid DNA), of which 60% contained class 1 integrons, 15% contained class 2 integrons, and 11.7% carried integrons of both classes. Thus, it may be concluded that integrons are the common mediators of antimicrobial resistance among commensal multidrug resistant Escherichia coli at important stages of poultry production.
Similar content being viewed by others

A novel antibiotic class targeting the lipopolysaccharide transporter
Claudia Zampaloni, Patrizio Mattei, … Kenneth A. Bradley

Molecular mechanisms of antibiotic resistance revisited
Elizabeth M. Darby, Eleftheria Trampari, … Jessica M. A. Blair

Antibiotic resistance in the environment
D. G. Joakim Larsson & Carl-Fredrik Flach
Introduction
The increasing resistance to commonly applied antimicrobial agents is being reflected by growing multiple drug resistance (MDR) in bacteria and is becoming a growing threat to public health. The use of antimicrobial agents in animal husbandry has been linked to the development and spread of the resistant bacteria 1 . Resistant bacteria can be transferred for example from poultry products to humans via consuming or handling meat contaminated with pathogens 2 . However, the resistance of commensal bacteria is equally important as they constitute a reservoir and vector of resistance determinants in the environment 3 .
Exposure to antimicrobials of different classes can lead to cross-resistance and the selection of antibiotic resistance genes (ARGs) that may spread laterally on mobile genetic elements (MGEs) via horizontal gene transfer (HGT) 4 . HGT is a phenomenon in which genes are transferred between organisms of either the same or different species which often remain in a close ecological relationship 5 .
It has been shown that conjugation is one of the key mechanisms responsible for the spread of the ARGs 6 . One of the most efficient mechanism of acquiring ARGs is facilitated by integrons—a site-specific recombination systems capable of recruiting open reading frames (ORF) in the form of mobile gene cassettes 7 . Integrons are divided into two distinct subsets, mobile integrons (MIs)—linked to mobile DNA elements, which are primarily involved in the spread of ARGs, and chromosomal integrons (CIs). MIs are associated with conjugation plasmids or transposons (the integron itself is not mobile), which allows the spread and exchange of the resistance genes between individual strains and bacterial species 8 . MIs may be divided into five classes, which are involved in the propagation of antibiotic-resistance genes 9 . These classes are divided based on the sequence of the encoded integrase genes, which share 40–58% sequence homology. The first three classes of integrons are involved in the acquisition of the MDR phenotype. Class 1 integrons are mostly found in clinical and animal production isolates, and most of the known antibiotic-resistance gene cassettes belong to this group. Class 1 is associated with functional and nonfunctional transposons derived from Tn402 that can be embedded in larger transposons, such as Tn21 10 . Class 2 integrons are associated with Tn 7 derivatives, and class 3 integrons are thought to be located in a transposon inserted in plasmids 11 , 12 . The integron types may be identified based on the detection of the specific integrase gene sequence— Int 1, Int 2 and Int 3 respectively 13 .
Class 1 and 2 integrons are frequently detected and well characterized, mostly among bacteria belonging to the Enterobacteriaceae family, including E. coli 14 . The majority of E. coli strains are commensals inhabiting the intestinal tract of humans and warm-blooded animals and rarely causes diseases 15 . The adaptation ability of these microorganisms into various niches in host organism is determined by its extremely plastic genome. Another important driving force for the evolution of the E. coli genome is the mobile gene pool replaced by the HGT 16 .
Studies of microorganisms found in the breeding environment of broiler chickens (including E. coli ), such as feces and poultry meat provide valuable information about the reservoir of bacterial genes 17 . While assessing the risks arising from the possible transfer of resistant bacteria within the poultry production chain, it seems important to know the diversity and the prevalence of genetic determinants of antibiotic resistance among the commensal E. coli strains.
In the recent years, a number of studies have been carried out aiming to identify the presence and the structure of the integrons, the type of the resistance cassettes and the relationship between the occurrence of integrons and MDR, in commensal and pathogenic E. coli isolated from human and animal samples 18 , 19 , 20 . A link between the use of antibiotics in animal production and antimicrobial resistance of human pathogens (within which food is one of the possible vectors) was reported in several studies 21 , 22 , 23 . Nevertheless, little is known about the distribution of integrons in E. coli isolated from commercial broiler meat in Poland. Therefore, the purpose of this study was to determine antimicrobial resistance profiles, distribution of class 1 and 2 integrons and integron-associated gene cassettes in commensal strains isolated from poultry litter, broiler chicken feces and meat in western Poland.
Antimicrobial resistance phenotypes of E. coli isolates
We have analyzed the incidence of multidrug resistance gene sequences and the prevalence of class 1 and 2 integrons within 74 commensal E. coli isolates, obtained from poultry litter (group A, n = 23), swabs from broiler chicken cloaca (group B, n = 26) and poultry meat (group C, n = 25). 60 (81.1%) of them exhibited a multiresistant phenotype (resistance to at least three different antimicrobial agent families). In the first step of study, the phenotypic resistance of the E. coli isolates to six antibiotics and chemotherapeutics was assessed.
Out of 23 E. coli isolates obtained from poultry litter, 16 showed multidrug resistance. The highest resistance was recorded for AMP (100%), DOX (100%), CIP and STX (81.3%), and AMC (75%); the lowest for GEN (12.5%). Out of 26 isolates obtained from chicken cloaca, 25 exhibited MDR. Among the examined MDR isolates, the highest percentage of resistance was observed for the following antibiotics: AMP (100%), CIP (92%), AMC, DOX (88%), STX (84%) and the lowest for GEN (36%). All E. coli isolates obtained from the cloaca of chickens, showed phenotypic resistance to the antibiotics classes with which the broilers were treated on farms. Among E. coli isolates obtained from meat, 19 of them showed MDR. In this group, the highest resistance was observed for AMP (100%). Lower resistance was noted for: STX (94.7%), CIP (78.9%), DOX (68.4%). Resistance to AMC and GEN was noted in 42.1% and 36.8% of E. coli isolates respectively. Figure 1 ., shows the percentage of phenotypic resistance to 6 antibiotics among MDR E. coli isolates obtained from poultry litter, cloacal swabs and poultry meat (group A, B and C). Antimicrobial susceptibility tests showed that 11.7% of all MDR isolates of E. coli , were resistant to three antibiotics, 38.3% to four, and 35% showed resistance to five and 15% to six antibiotics (Table 1 ). Among all E. coli isolates positive for integron sequences, the most common drug resistance profile was that of the resistance to: AMP, AMC, CIP, STX, DOX. In the case of 2 isolates with a pair of integrons, E. coli isolates that showed resistance to 6 antimicrobial agents were reported. Their resistance profile was the same: AMP, AMC, CIP, GEN, STX, DOX.
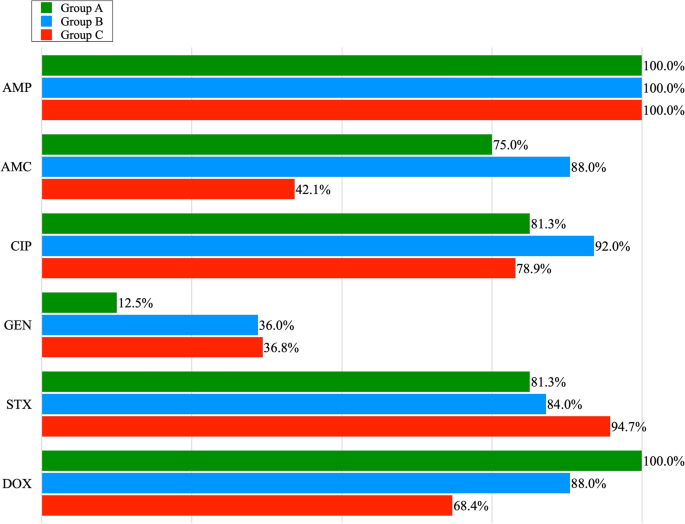
The percentage of phenotypic resistance to 6 antibiotics among the MDR E. coli isolates obtained from poultry litter (Group A), cloacal swabs (Group B) and poultry meat (Group C): AMP—Ampicillin; AMC—Amoxycyline with Clavulanic Acid; CIP—Ciprofloxacin; GEN -Gentamicine; STX—Sulfamethoxazole with Trimetoprime; DOX—Doxycycline.
Identification and characteristics of resistance genes.
Resistance to AMP and AMC encoded by the narrow spectrum beta lactamase resistance gene ( bla TEM ) was found in genomic DNA of all E. coli isolates from groups A and B, and in 63,2% of the poultry meat (group C). In case of other genes encoding beta-lactamase resistance, the occurrence of bla CTX-M gene was noted in one colon isolate. Bla SHV gene was detected in one isolate from the poultry meat swabs. Among the MDR isolates showing the ciprofloxacin (CIP) resistance phenotype, a total of 7 qnrA genes, 10 qnrB genes and 6 qnrS genes were reported in genomic DNA. 18 MDR isolates were gentamicin-resistant, and the strA-strB , aphA1 , and aac(3)-II genes, giving resistance to aminoglycosides, were present in: 13 isolates of E. coli obtained from litter, 19 isolates from cloacal swabs and 5 isolates from poultry meat. 13 of the 16 tested E. coli MDR isolates showed a sulphonamides resistance phenotype which was encoded by: sul1 (50%), sul2 (18.8%) and sul3 (25%). In the case of 21 MDR isolates obtained from cloaca the sul1 and sul3 genes were recorded in 28% and the sul 2 gene in 44% of the cases. In this group, the presence of pairs of sul genes was noted in 5 isolates, in the su1—sul2 combination. In the MDR group of meat isolates, 18 E. coli isolates contained the following genes: sul 1 (31.6%), sul 2 (26.3%) and sul3 (10.5%) and in one case a pair of genes ( sul1 and sul2 ) were noted. In the case of E. coli isolates recovered from the litter, one of them confirmed 3 genes ( sul1, sul2, sul3 ) determining resistance to sulfonamides. Sulfonamide-resistant isolates of E. coli showed in most cases compatible resistance to trimethoprim resistance genes ( dfr1, dfr5, dfr7/17 ) in 84.6% cases in group A, 71.4% in group B and 38.9% in group C. The tet genes ( tetA, tetB ) giving resistance to doxycycline were found in 75 and 37.5% isolates from poultry liter, 52 and 36% isolates from cloacal swab, and 36.8 and 10.5% isolates from meat. The tetC gene was not found in any of the studied groups. The pairs of tetA and tetB genes were found in groups A and B in 4 and 3 cases, respectively. The prevalence of the resistance genes among multidrug resistant E. coli isolates obtained from poultry litter (Group A), cloacal swabs (Group B) and poultry meat (Group C) is shown in Table 2 and Fig. 2 .
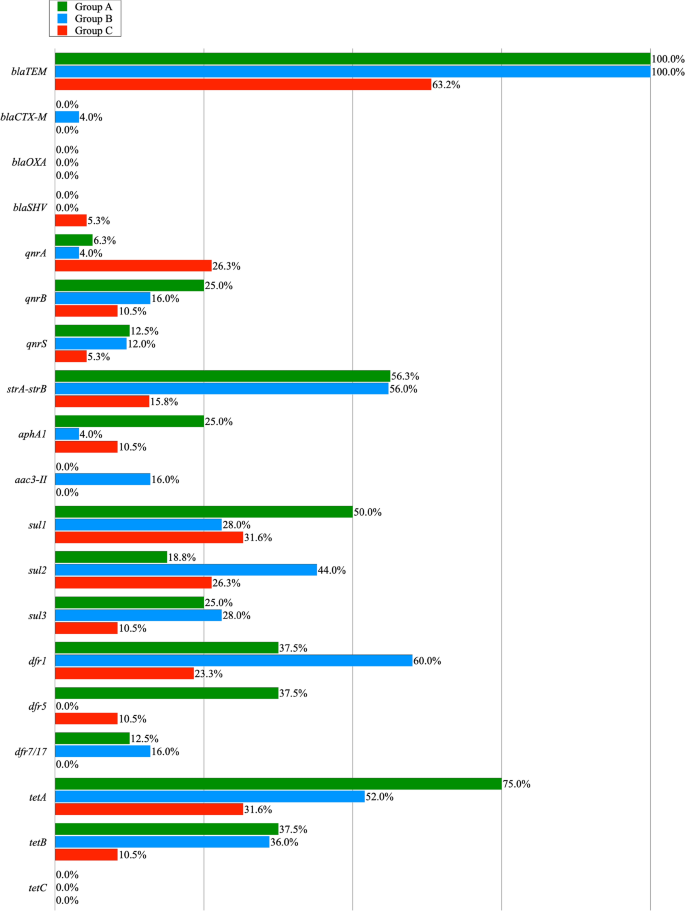
The prevalence of resistance genes among the MRD resistant E. coli isolates obtained from poultry litter (Group A), cloacal swabs (Group B) and poultry meat (Group C).
Detection and characterization of integrons
Our study reports a high incidence of integron-bearing E. coli isolates. More than half of the multidrug resistant isolates contained integrons. The presence of class 1 and 2 integrons was confirmed in 45 out of 60 MDR E. coli isolates (75%). Structure of 1 class integron, identified by the presence of the IntI1 gene was detected in 36 (60%) multiresistant isolates in plasmid DNA. Most of this gene was recorded in the DNA of bacteria isolated from cloaca—15 E. coli isolates, and next from litter and meat—11 and 10, isolates, respectively. The frequency of the integrons of class 1 was not significantly different across sampling locations ( P > 0.133). In turn, class 2 integrons, identified by the presence of the intI2 gene, were found in a much smaller number of cases, only in cloacal and meat isolates (4 and 5 cases). The frequency of the integtons of class 2 differed significantly ( P ≤ 0.05) between these two locations. In the samples in which both classes of the integrons were detected (group B and C), the class 1 integrons were significantly more frequent than class 2 integrons (B: P ≤ 0.01, and C: P ≤ 0.05).
Gene cassette analysis of class 1 and 2 integron genes
Table 3 , 4 and 5 shows the phenotypes, antibiotic resistance, and prevalence of class 1 and 2 integrons and their resistance gene cassettes in MDR E. coli isolates obtained from all groups. E. coli isolates containing integrons were grown from poultry litter (group A). 11 out of 16 MDR isolates, contained class 1 integrons only. The variable region of class 1 integrons most frequently contained the aadA 1 gene cassette in seven cases. The dfrA1-aadA1 cassette series was present in three cases (18.8%) and dfrA17-aadA5 in one strain. In the class 2 integron variable region, no gene cassettes were found in any case (Table 3 ).
In case of the multidrug-resistant E. coli isolates obtained from cloacal swabs from broiler chickens (group B), 15 isolates contained class 1 integrons and 4 isolates contained class 2 integrons (Table 4 ). The results of DNA sequencing of the inserted gene cassettes allowed to identify 11 class 1 integrons containing 5 different gene cassettes: aadA1 (36.4%), dfrA5 (18.2%) and arrays of cassettes: dfrA1-aadA1 (36.4%) and dfrA17-aadA5 (9.1%) in five separate isolates. Genes contained in the four class 2 integron cassettes contained gene cassette arrays: dfrA1-sat2-aadA1 .Two isolates containing class 1 and 2 integrons contained cassette arrays: aadA1; dfrA1-sat2-aadA1 and dfrA5; dfrA1-sat2-aadA1 . Four (26.7%) class 1 positive integron isolates had no gene cassettes in their variable part.
In the group of MDR E. coli isolated from the poultry meat (group C), genes contained in the class 1 integron variable region were detected in 9 cases (Table 5 ). VR class 1 integrons showed less variation compared to group B isolates and usually contained 2 gene cassette arrays: aadA1 (22.2%) and dfrA1-aadA1 (77.8%). The variable region of 5 class 2 integrons contained in only two cases a set of cassettes: dfrA1-sat2-aadA1 . In one strain, a pair of class 1 and 2 integrons was detected as well as their variable parts were empty.
It is estimated that in most developed nations, livestock use 50–80% of antibiotics produced 24 . Commonly used groups of veterinary medicinal products in the EU are tetracyclines (32%), penicillins (26%), sulfonamides (12%), macrolides (7%), polymyxins (5%) and aminoglycosides (5%) 25 , 26 .
In Poland, 829 tones of antibiotics are used annually, of which as much as 578 tones in the agricultural industry 27 , 28 . The most commonly used groups of substances in years 2014–2016 were tetracyclines (42.34–49.07%), penicillins (18.98–23.40%) and macrolides (11.69–13.22%) 26 .
In our study, 74 unrelated commensal isolates of Escherichia coli originated from poultry litter, cloacal swabs and poultry meat were phenotypically and genotypically tested for the antimicrobial resistance and the presence of integrons as factor, for the development of antibiotic resistance and the emergence of MDR strains. Among them, 60 isolates (81.1%) were multiresistant (resistant to a minimum of three classes of antibiotics). We have found the highest level of antimicrobial resistance (96.2%) in the E. coli isolates obtained from broiler intestinal swabs (group B).
The high incidence of multidrug resistance in our study, particularly regarding isolates obtained from feces and meat, is extremely significant and should be regarded as a serious health risk due to the fact, that multidrug resistant isolates may have a chance of contaminating food products, and consequently being transferred to humans.
The percentage of resistance to some antimicrobial agents (Ampicillin, Doxycycline, Trimetophrim, Sulfamethoxazole, and Ciprofloxacin) in all studied groups was particularly high (100–68.4%), which indicates that the commensal E. coli isolates may be a reservoir of resistance to antibiotics and chemotherapeutics. Our data largely overlaps with the data made available by the European Food Safety Authority and the European Centre for Disease Prevention and Control, on the resistance profile of the commensal E. coli isolates obtained from slaughterhouse broilers, collected between 2009 and 2014 in Poland 28 . It confirms high resistance of E. coli isolates to nalidixic acid ciprofloxacin, and ampicillin (70–90%) and a limited resistance to tetracyclines, sulfonamides and streptomycin 28 , 29 .
The bla TEM gene encoding β lactamase, which gives resistance to penicillins and cephalosporins of the first generation, has been detected in all multidrug-resistant isolates obtained from litter and feces. The dominance of the TEM gene over the CTX gene in fecal poultry isolates was also noted in the Nigerian 30 study but was not as pronounced as in our experiment (63%- TEM and, 35% CTX-M ). Although E. coli isolates with enzymes belonging to the CTX-M family in ESBL-positive bacteria are currently predominant in the world 31 in our study the bla CTX-M gene was reported only in one bacterial strain (4%) from cloacal swabs. Similar results were obtained in China, where bla CTX-M was detected in 1.6% of the isolates coming from the meat 32 .
Quinolone resistance is a current worldwide problem in human and veterinary medicine 33 . Quinolone resistance can encoded in bacterial chromosome or be present in plasmids. Plasmid-mediated quinolone resistance (PMQR) promote the spread of the multi-drug resistance phenotype. For example, qnr genes present on MDR plasmids are often found with genes encoding β-lactamases 34 . Of all Qnr determinants present in the our study, the qnrB gene was found most frequently. Similar results were published in other studies 33 , 35 . The occurrence of PMQR is also associated with resistance to other groups of antibiotics. The mechanism responsible for this phenomenon is related to the presence of the aminoglycoside acetyltransferase enzyme—AAC(6')-Ib-cr, which modifies both the molecular structure of some fluoroquinolones and aminoglycosides or the oqxAB gene encoding an MDR-type efflux pump contributing to increased resistance to quinolones and chloramphenicol, trimethoprim and quinolones 36 , 37 . In Seo and Lee 33 study, 10 PMQR-positive E. coli were isolated from chicken meat, and these isolates also showed higher resistance rates to several antimicrobial agents when compared to PMQR-negative E. coli . This is consistent with previous studies showing that the PMQR genes increase resistance to other antimicrobials and cause MDR to drugs such as: aminoglycosides, β-lactams, chloramphenicol, sulfonamides, tetracyclines and trimethoprim 38 .
Tetracyclines are commonly used to treat bacterial infections in livestock, including poultry in many countries 39 , 40 , 41 . Due to the numerous advantages of tetracyclines, such as their widespread availability, low cost, and several side effects, the use of such antibiotics to treat animal and human infections has been increasing in recent years 42 . The chickens are treated with tetracycline orally and their metabolites (up to 90%) are excreted in the feces 43 on manure 44 . It is noteworthy that in our study the highest resistance to doxycycline was recorded among E. coli isolates derived from poultry litter, which is a mixture of poultry manure, litter, feathers, feed, and spilled drinking water that accumulates during breeding 45 . However, the proportion of E. coli isolates with resistance to tetracyclines was lower than the proportion of E. coli isolates resistant to beta lactam antibiotics. Similar results were obtained in study by Islam et al. 46 in which MDR isolates were the most resistant to tetracyclines (96.6%) and penicillins (100%). In addition to the antibiotic residues, manure also contains MDR bacteria and resistance genes, which can be transmitted to humans through direct contact between poultry and humans or indirectly via the food chain 47 . The results of genotyping showed that similarly to other published data, the resistance of commensal E. coli to tetracyclines was induced by the presence of tetA and tetB genes 48 . The highest content of tet genes in poultry litter isolates confirms the thesis put forward by Furtula et al. 44 , that the breeding environment significantly contributes to the spread of the resistance, via the transmission of the resistance genes.
Resistance to sulfonamides in Gram-negative bacteria generally results from the presence of the genes sul1 , sul2 , and/or sul3 . Among them, the sul2 gene is the most widely distributed sul gene in porcine, avian, or human E. coli isolates, and it plays an important role in sulfonamide resistance 49 . In our research, the prevalence of sul2 genes was highest in isolates from cloaca swabs (44%) and was similar to the results of other studies 50 , 51 . Interestingly, in only one strain obtained from litter, all tested sul genes were determined. The frequency of sul genes detection in our experiment corresponded to other studies 50 , 51 . The selective pressure exerted by sulfonamides in the poultry industry appears to be high, which may favor the maintenance of acquired sul genes among bacteria 52 .
In our study, the total prevalence of integrons (75%) in MDR isolates was higher than the prevalence of integrons detected in other poultry production prevalence studies 53 , 54 . We also found a clear predominance of class 1 integrons in relation to class 2 integrons, which is consistent with previous studies that also showed the highest prevalence of this class in poultry isolates 55 , 56 .
Most of the integrons detected in our study contained gene cassettes encoding resistance to trimethoprim ( dfrA gene type) and streptomycin/spectinomycin belonging to aminoglycosides ( aadA gene type), and the most frequently detected sequence of cassettes was dfrA1-aadA1 . The persistence of these genes, which have been reported worldwide in isolates from different sources, may be related to the widespread use of streptomycin/spectinomycin, trimethoprim, sulfonamides and other antibiotics in food producing animals. Although the afore-mentioned aminoglycosides are not used therapeutically in animals in Poland, the presence of aadA genes may be a form of genetic memory, in case of re-exposure of the microorganism to this group of antibiotics 57 .
The analysis of the variable part of integrons in our experiments indicated the presence of one to three gene cassettes. In group B, we noted the highest number of integrons among all tested groups and a greater variety of gene cassettes within the integron variable part. The higher prevalence of class 1 integrons among the E. coli isolates obtained from the cloaca may be caused by the development and spread of the resistance genes, due to the misuse or abuse of antibiotics in the poultry production 1 .
Of all MDR E. coli isolates that had integrons, 8 isolates (13.3%) did not contain any of the evaluated gene cassettes. The situation regarding the so-called "empty integrons" has already been described by Fonseca et al. 58 , where it was indicated that these bacteria could rapidly develop into MDR in the future. However, it cannot be ruled out that integrons may have previously removed cassettes of resistance genes acquired by cutting them out for unknown reasons 59 .
The data obtained in this study highlights the importance of commensal E. coli in the spread of resistance genes at different stages of poultry production. We confirmed that more than half of the multidrug-resistant isolates (75%) contained integrons. Furthermore, we showed that antibiotic resistance can also occur on non-integron structures. Therefore, there is a need for further detailed genetic studies on the evolution of isolates present in poultry to uncover the underlying mechanisms the acquisition of resistance by these microorganisms and to analyze the implications for humans. Such data may be used to determine the dynamics of resistance development and strategies to counteract antibiotic resistance among zoonotic microorganisms transmitted through food of animal origin at all stages of the food chain, from farm to table.
Materials and methods
E. coli isolates.
A total number of 74 E. coli isolates was collected from three areas of poultry production: litter swabs from chicken houses (n = 23), cloacal swabs from chicken (n = 26), chicken meat from slaughterhouses (n = 25). All samples were collected between November 2019 and March 2020.
Litter samples from 23 different chicken houses were acquired in accordance with the boot swabs sampling procedure guidelines of the national control program for Salmonella serotypes in poultry flocks in line with the guidelines of the current EU law 60 . Samples were collected from 4-week-old chicks (average weight 1.6 kg) 2 weeks before slaughter. The samples of cloacal swabs were collected using swabs (NRS II Transwab swabs with 10 Buffered Peptone Water, Medical Wire & Equipment, Corsham, United Kingdom) in a poultry slaughterhouse. Chickens were raised on 25 unrelated farms located in Greater Poland Voivodship. Birds that were sent to slaughter at 6 weeks of age, weighed an average of 3 kg and belonged to the Ross 308 breed. The chicken meat samples were obtained from neck skin. These samples were delivered to the laboratory for testing, as part Salmonella monitoring program, from 5 different poultry slaughterhouses located in the Greater Poland Voivodeship, from different periods of production.
Information regarding the antibiotics used in the above chickens (name of the antibiotic, withdrawal periods) was included in the food chain documentation. In the treatment of poultry, the most frequently used antibiotics were: Amoxicillin, Enrofloxacin, Doxycycline, Sulfamethoxazole / Trimethoprim.
The samples were placed in buffered peptone water (BioMerieux, Marcy l'Etoile, France) and incubated at 35 °C (± 1 °C) for 18 h (± 2 h) under aerobic conditions. Next, the material was plated on MacConkey agar medium (OXOID, Basingstoke, United Kingdom) and incubated for 24 ± 2 h under aerobic conditions at 37 °C ± 1 °C. Colonies with the typical E. coli phenotype were selected (one per sample) and verified by the MALDI-TOF method (Bruker, Bremen, Germany). The score ranged from 2.015 to 2.152.
Antimicrobial susceptibility testing
Antibiotic susceptibility tests of all 74 E. coli isolates were performed following the standard agar disk diffusion method, according to the CLSI (Clinical and Laboratory Standards Institute-2012) using commercially available antimicrobial disks containing (OXOID, Basingstoke, United Kingdom): penicillins (Ampicillin—AMP 10 μg and Amoxycyline with Clavulanic acid—AMC 20/10 μg), fluoroquinolones (Ciprofloxacin—CIP 5 μg), aminoglycosides (Gentamicine—GEN 10 μg), sulfonamides (Sulfamethoxazole with Trimetoprime—STX 25 μg), tetracyclines (Doxycycline—DOX 30 μg).
The following media were used for the tests: Mueller Hinton Broth (Thermo Fisher Scientific, Waltham, Massachusetts, USA), Mueller–Hinton agar (OXOID, Basingstoke, United Kingdom). The bacterial colonies were classified as sensitive, intermediate, or resistant according to the standardized CLSI guidelines (VET 01S—Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals) and E. coli ATCC 25922 strain was used as control. The isolates were collected and stored at -80℃ for further analyses on Viabank medium (OXOID, Basingstoke, United Kingdom).
Detection of integrons
Class 1 and Class 2 integrons were detected based on the presence of gene sequences characteristic for integrase 1 (IntI1 ) and integrase 2 ( IntI2 ) respectively. Selected regions were amplified by qualitative PCR carried out on plasmid DNA extracted from the three studied groups (poultry litter, feces and carcasses).
Plasmid DNA from the bacterial samples was extracted with the Gene Matrix Plasmid Miniprep DNA Purification Kit (E3500 EurX, Gdansk, Poland) according to the manufacturer’s protocol. PCR amplification was done in a 10µL mixture containing: 1µL DNA template, 0.3µL of primers (0.3 μM), 2µL of 5× HOT FIREPol® Blend Master Mix kit (04-25-00S25, SolisBiodyne, Tartu, Estonia) and molecular biology graded water (nuclease free, W4502 Merck, Darmstadt, Germany). The thermal cycling conditions included: preincubation at 95 °C (15 min), followed by 38 cycles of denaturation at 95 °C (20 s), annealing at 61 °C (45 s), extension at 72 °C (60 s) and final extension at 72 °C (5 min). Primer pair sequences are listed in Table 6 . The specificity of the PCR reaction (product length–base pair, bp) was verified by electrophoresis on a 1.5% agarose gel.
Sequencing of the variable regions of Integron 1 and Integron 2
In the bacterial isolates containing the IntI1 and the IntI2 genes, the variable regions (VRs) of both of the studied integrons were sequenced in order to reveal the specific DNA sequence (plausibly bearing the multidrug resistance genes) within the integron structure. The regions were amplified with a set of specific primers: Integron CL (for integron 1) and Integron CL JJ (for integron 2), sequences are listed in Table 6 . The PCR reaction conditions, and primer concentrations were the same as described for IntI1 and IntI2 amplification, with an annealing temperature of 61 °C. The amplified gene cassettes of similar PCR product length (base pair, bp) were sequenced by the Sanger method. Prior to sequencing, the PCR products were purified with FastAP and ExoI enzymes (EF0654 and EN0581, Thermo Fisher Scientific, Waltham, Massachusetts, USA), amplified With the BigDye™ terminator v3.1 Cycle Sequencing Kit (4,337,458, Life Technologies, Carlsbad, California, USA) and purified on Sephadex G50 (G5050, Sigma, St. Louis, Missouri, USA) by filtration. Sequencing of the cassette arrays was done with Applied Biosystems ABI 3130xl 16-capillary array genetic analyzer (Applied Biosystems, Waltham, Massachusetts, USA). Data was analysed with the Seqman software.
Detection of antimicrobial resistance genes outside the integron cassettes
This experiment focused on the detection of 19 antimicrobial resistance genes in the studied genomic samples. Genomic DNA was extracted with Extractme DNA Bacteria Kit (EM02, Blirt, Gdansk, Poland) according to the manufacturer’s protocol. The PCR reaction conditions were the same as described for the detection of introns. Primer sequences are listed in Table 6 . We tested for the presence of 4 genes associated with resistance to b-lactam antibiotics—ampicillin and amoxicillin ( bla SHV , bla TEM , bla CTX-M , bla OXA ) 61 , ciprofloxacin-resistance ( qnrA, qnrB, qnrS ) 62 , streptomycin-resistance ( strA-strB) 63 , kanamycin-resistance ( aphA1 ) 63 , gentamicin-resistance ( aac(3)-II ) 64 , sulphonamides-resistance ( sul1, sul2, sul3 ) 63 , trimethoprim-resistantance ( dfr1, dfr5, dfr7/17 ) 65 and tetracycline-resistance ( tetA, tetB, tetC ) 63 . The analyses were done with the set of primers listed in Table 6 and the PCR reaction conditions were as described for the detection of integrons.
Statistical analysis
The significance of the differences between presence and absence of integrons of class 1 and 2, and the predominance of class 1 integrons in relation to class 2 integrons were tested within each group (A, B, and C) using Pearson's Chi-squared Test. Relation between the presence of integrons and the phenotype of multi antibiotic resistance was tested with the use of Poisson Regression analyses where the dependent variable was the number of antibiotics, resistance, and the independent variable was the integron’s presence. These analyses were performed separately for integron class 1 and 2 and for each group (A, B, and C). All of the statistical analyses were performed with the use of the R environment 66 .
Agyare, C., Boamah, V. E., Zumbi, C. N. & Osei, F. B. Antibiotic Use in Poultry Production and Its Effects on Bacterial Resistance. https://doi.org/10.5772/intechopen.79371 (2018).
Article Google Scholar
van den Bogaard, A. E. & Stobberingh, E. E. Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14 , 327–335. https://doi.org/10.1016/s0924-8579(00)00145-x (2000).
Article PubMed Google Scholar
Racewicz, P., Majewski, M., Madeja, Z. E., Lukomska, A. & Kubiak, M. Role of integrons in the proliferation of multiple drug resistance in selected bacteria occurring in poultry production. Br Poult Sci 61 , 122–131. https://doi.org/10.1080/00071668.2019.1697426 (2020).
Article CAS PubMed Google Scholar
Skarżyńska, M., Zając, M. & Wasyl, D. Antibiotics and bacteria: Mechanisms of action and resistance strategies. Postępy Mikrobiologii - Advancements of Microbiology 59 , 49–62. https://doi.org/10.21307/pm-2020.59.1.005 (2020).
Kelly, B. G., Vespermann, A. & Bolton, D. J. The role of horizontal gene transfer in the evolution of selected foodborne bacterial pathogens. Food Chem. Toxicol. 47 , 951–968. https://doi.org/10.1016/j.fct.2008.02.006 (2009).
Singer, R. S. & Williams-Nguyen, J. Human health impacts of antibiotic use in agriculture: A push for improved causal inference. Curr. Opin. Microbiol. 19 , 1–8. https://doi.org/10.1016/j.mib.2014.05.014 (2014).
Stokes, H. W. & Hall, R. M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3 , 1669–1683. https://doi.org/10.1111/j.1365-2958.1989.tb00153.x (1989).
Boucher, Y. et al. Recovery and evolutionary analysis of complete integron gene cassette arrays from Vibrio. BMC Evolu. Biol. https://doi.org/10.1186/1471-2148-6-3 (2006).
Ghazalibina, M. et al. Prevalence of integrons and antibiotic resistance pattern in Acinetobacter baumannii isolated from clinical samples of Iranian patients: A systematic review and meta-analysis. Ethiop. J. Health Sci. 29 , 639–648. https://doi.org/10.4314/ejhs.v29i5.15 (2019).
Article PubMed PubMed Central Google Scholar
Cambray, G., Guerout, A. M. & Mazel, D. Integrons. Annu. Rev. Genet. 44 , 141–166. https://doi.org/10.1146/annurev-genet-102209-163504 (2010).
Barlow, R. S., Pemberton, J. M., Desmarchelier, P. M. & Gobius, K. S. Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob. Agents Chemother. 48 , 838–842. https://doi.org/10.1128/aac.48.3.838-842.2004 (2004).
Article CAS PubMed PubMed Central Google Scholar
Shibata, N. et al. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J. Clin. Microbiol. 41 , 5407–5413. https://doi.org/10.1128/jcm.41.12.5407-5413.2003 (2003).
Kargar, M., Mohammadalipour, Z., Doosti, A., Lorzadeh, S. & Japoni-Nejad, A. High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in Southwest of Iran. Osong. Public Health Res. Perspect. 5 , 193–198. https://doi.org/10.1016/j.phrp.2014.06.003 (2014).
Han, N., Sheng, D. & Xu, H. Role of Escherichia coli strain subgroups, integrons, and integron-associated gene cassettes in dissemination of antimicrobial resistance in aquatic environments of Jinan, China. Water Sci. Technol. 66 , 2385–2392. https://doi.org/10.2166/wst.2012.473 (2012).
Ramos, S. et al. Escherichia coli as commensal and pathogenic bacteria among food-producing animals: Health Implications of Extended Spectrum beta-lactamase (ESBL) Production. Animals (Basel). https://doi.org/10.3390/ani10122239 (2020).
Mazurek, J., Bok, E., Stosik, M. & Baldy-Chudzik, K. Antimicrobial resistance in commensal Escherichia coli from pigs during metaphylactic trimethoprim and sulfamethoxazole treatment and in the post-exposure period. Int. J. Environ. Res. Public Health 12 , 2150–2163. https://doi.org/10.3390/ijerph120202150 (2015).
Garcia-Aljaro, C., Moreno, E., Andreu, A., Prats, G. & Blanch, A. R. Phylogroups, virulence determinants and antimicrobial resistance in stx(2) gene-carrying Escherichia coli isolated from aquatic environments. Res. Microbiol. 160 , 585–591. https://doi.org/10.1016/j.resmic.2009.08.004 (2009).
Drugdova, Z. & Kmet, V. Prevalence of beta-lactam and fluoroquinolone resistance, and virulence factors in Escherichia coli isolated from chickens in Slovakia. Biologia 68 , 11–17. https://doi.org/10.2478/s11756-012-0142-6 (2013).
Article CAS Google Scholar
Kaushik, M., Kumar, S., Kapoor, R. K., Virdi, J. S. & Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 51 , 167–176. https://doi.org/10.1016/j.ijantimicag.2017.10.004 (2018).
Seo, K. W. & Lee, Y. J. Prevalence and characterization of beta-lactamases genes and class 1 integrons in multidrug-resistant Escherichia coli isolates from chicken meat in Korea. Microb. Drug Resist. https://doi.org/10.1089/mdr.2018.0019 (2018).
Stine, O. C. et al. Widespread distribution of tetracycline resistance genes in a confined animal feeding facility. Int. J. Antimicrob. Agents 29 , 348–352. https://doi.org/10.1016/j.ijantimicag.2006.11.015 (2007).
Article ADS CAS PubMed Google Scholar
Carattoli, A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14 , 117–123. https://doi.org/10.1111/j.1469-0691.2007.01851.x (2008).
Silbergeld, E. K., Graham, J. & Price, L. B. Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 29 , 151–169. https://doi.org/10.1146/annurev.publhealth.29.020907.090904 (2008).
Cully, M. Public health: The politics of antibiotics. Nature 509 , S16-17. https://doi.org/10.1038/509S16a (2014).
EMA. Sales of veterinary antimicrobial agents in 30 European countries in 2016. (European Surveillance of Veterinary Antimicrobial Consumption, EMA/275982/2018, 2018).
Majewski, M., Anusz, K., Belkot, Z., Racewicz, P. & Lukomska, A. Impact of residues of veterinary medicinal products in food of animal origin on public health safety in Poland in the years 2003–2017. Med. Weter. 76 , 416–422. https://doi.org/10.21521/mw.6412 (2020).
ECDC, EFSA & EMA. Second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. https://doi.org/10.2903/j.efsa.2017.4872 (2017).
EFSA. European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. Efsa J. https://doi.org/10.2903/j.efsa.2017.4694 (2017).
Roth, N. et al. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli : A global overview. Poult. Sci. 98 , 1791–1804. https://doi.org/10.3382/ps/pey539 (2019).
Akinbami, O. R., Olofinsae, S. & Ayeni, F. A. Prevalence of extended spectrum beta lactamase and plasmid mediated quinolone resistant genes in strains of Klebsiella pneumonia , Morganella morganii , Leclercia adecarboxylata and Citrobacter freundii isolated from poultry in South Western Nigeria. PeerJ https://doi.org/10.7717/peerj.5053 (2018).
Saliu, E. M., Vahjen, W. & Zentek, J. Types and prevalence of extended-spectrum beta-lactamase producing Enterobacteriaceae in poultry. Anim. Health Res. Rev. 18 , 46–57. https://doi.org/10.1017/s1466252317000020 (2017).
Li, L. L., Ye, L., Kromann, S. & Meng, H. C. Occurrence of extended-spectrum beta-lactamases, plasmid-mediated quinolone resistance, and disinfectant resistance genes in Escherichia coli isolated from ready-to-eat meat products. Foodborne Pathog. Dis. 14 , 109–115. https://doi.org/10.1089/fpd.2016.2191 (2017).
Seo, K. W. & Lee, Y. J. Prevalence and characterization of plasmid mediated quinolone resistance genes and class 1 integrons among multidrug-resistant Escherichia coli isolates from chicken meat. J. Appl. Poultry Res. 28 , 761–770. https://doi.org/10.3382/japr/pfz016 (2019).
Piekarska, K. et al. Prevalence of qnr genes in clinical Enterobacteriaceae non-susceptible to fluoroquinolone in Poland. Med. Dosw. Mikrobiol. 64 , 211–219 (2012).
CAS PubMed Google Scholar
Kindle, P. et al. Phenotypic and genotypic characteristics of Escherichia coli with non-susceptibility to quinolones isolated from environmental samples on pig farms. Porcine Health Manag. 5 , 9. https://doi.org/10.1186/s40813-019-0116-y (2019).
Robicsek, A. et al. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12 , 83–88. https://doi.org/10.1038/nm1347 (2006).
Hansen, L. H., Jensen, L. B., Sorensen, H. I. & Sorensen, S. J. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 60 , 145–147. https://doi.org/10.1093/jac/dkm167 (2007).
Amin, M., Dibachi, S., Shahin, M. Prevalence of class 1 integrons and plasmid-mediated qnr-genes among Enterobacter isolates obtained from hospitalized patients in Ahvaz, Iran. Infezioni Medicina , 351–357 (2017).
Sarmah, A. K., Meyer, M. T. & Boxall, A. B. A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65 , 725–759. https://doi.org/10.1016/j.chemosphere.2006.03.026 (2006).
Daghrir, R. & Drogui, P. Tetracycline antibiotics in the environment: A review. Environ. Chem. Lett. 11 , 209–227. https://doi.org/10.1007/s10311-013-0404-8 (2013).
Borghi, A. A. & Palma, M. A. Tetracycline: Production, waste treatment and environmental impact assessment. Braz. J. Pharm. Sci. 50 , 25–40. https://doi.org/10.1590/s1984-82502011000100003 (2014).
Jahantigh, M., Samadi, K., Dizaji, R. E. & Salari, S. Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran. BMC Vet. Res. https://doi.org/10.1186/s12917-020-02488-z (2020).
Kietzmann, M. & Baumer, W. Oral medication via feed and water - pharmacological aspects. Deutsche Tierarztliche Wochenschrift 116 , 204–208. https://doi.org/10.2376/0341-6593-116-204 (2009).
Furtula, V. et al. Veterinary pharmaceuticals and antibiotic resistance of Escherichia coli isolates in poultry litter from commercial farms and controlled feeding trials. Poult. Sci. 89 , 180–188. https://doi.org/10.3382/ps.2009-00198 (2010).
Font-Palma, C. Characterisation, kinetics and modelling of gasification of poultry manure and litter: An overview. Energy Convers. Manage. 53 , 92–98. https://doi.org/10.1016/j.enconman.2011.08.017 (2012).
Islam M.J., S. S., Das K.K., Sharmin N. and Hasan M.N. Isolation of Plasmid- Mediated Multidrug Resistant Escherichia coli from Poultry. Int. J. Sustain. Crop Prod. 3 , 46–50 (2008).
Marshall, B. M. & Levy, S. B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 24 , 718. https://doi.org/10.1128/cmr.00002-11 (2011).
Van, T. T. H., Chin, J., Chapman, T., Tran, L. T. & Coloe, P. J. Safety of raw meat and shellfish in Vietnam: An analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 124 , 217–223. https://doi.org/10.1016/j.ijfoodmicro.2008.03.029 (2008).
Blahna, M. T. et al. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J. Antimicrob. Chemother. 57 , 666–672. https://doi.org/10.1093/jac/dkl020 (2006).
Jouini, A. et al. Detection of multiple-antimicrobial resistance and characterization of the implicated genes in Escherichia coli isolates from foods of animal origin in Tunis. J. Food Prot. 72 , 1082–1088. https://doi.org/10.4315/0362-028x-72.5.1082 (2009).
Vinue, L. et al. Genetic environment of sul genes and characterisation of integrons in Escherichia coli isolates of blood origin in a Spanish hospital. Int. J. Antimicrob. Agents 35 , 492–496. https://doi.org/10.1016/j.ijantimicag.2010.01.012 (2010).
Soufi, L. et al. Escherichia coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Int. J. Food Microbiol. 144 , 497–502. https://doi.org/10.1016/j.ijfoodmicro.2010.11.008 (2011).
Adelowo, O. O., Fagade, O. E. & Agerso, Y. Antibiotic resistance and resistance genes in Escherichia coli from poultry farms, southwest Nigeria. J. Infect. Dev. Ctries. 8 , 1103–1112. https://doi.org/10.3855/jidc.4222 (2014).
Cavicchio, L. et al. Class 1 and class 2 integrons in avian pathogenic Escherichia coli from poultry in Italy. Poult. Sci. 94 , 1202–1208. https://doi.org/10.3382/ps/pev095 (2015).
Kang, H. Y. et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J. Antimicrob.. Chemother. 55 , 639–644. https://doi.org/10.1093/jac/dki076 (2005).
Dakic, I., Petrikkos, G., Dimitrijevic, V. & Charvalos, E. Multidrug resistance and integrons in Escherichia coli isolated from chicken in greece. Acta Vet.-Beogr. 61 , 575–584. https://doi.org/10.2298/avb1106575d (2011).
White, P. A., McIver, C. J. & Rawlinson, W. D. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob. Agents Chemother. 45 , 2658–2661. https://doi.org/10.1128/aac.45.9.2658-2661.2001 (2001).
Fonseca, E. L., Vieira, V. V., Cipriano, R. & Vicente, A. C. P. Class 1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol. Med. Microbiol. 44 , 303–309. https://doi.org/10.1016/j.femsim.2005.01.004 (2005).
Dessie, H. K., Bae, D. H. & Lee, Y. J. Characterization of integrons and their cassettes in Escherichia coli and Salmonella isolates from poultry in Korea. Poult. Sci. 92 , 3036–3043. https://doi.org/10.3382/ps.2013-03312 (2013).
EU. Commission Regulation (EU) No 200/2012 of 8 March 2012 concerning a Union target for the reduction of Salmonella enteritidis and Salmonella typhimurium in flocks of broilers, as provided for in Regulation (EC) No 2160/2003 of the European Parliament and of the Council Text with EEA relevance. In: Regulation, C. (Ed.) https://data.europa.eu/eli/reg/2012/200/oj (2012).
Fang, H., Ataker, F., Hedin, G. & Dornbusch, K. Molecular epidemiology of extended-spectrum beta-lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J. Clin. Microbiol. 46 , 707–712. https://doi.org/10.1128/jcm.01943-07 (2008).
Marti, E. & Balcazar, J. L. Real-time PCR assays for quantification of qnr genes in environmental water samples and chicken feces. Appl. Environ. Microbiol. 79 , 1743–1745. https://doi.org/10.1128/aem.03409-12 (2013).
Article ADS CAS PubMed PubMed Central Google Scholar
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J. & Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in Natural Environments in Ontario, Canada. Appl. Environ. Microbiol. 75 , 559–566. https://doi.org/10.1128/aem.01821-08 (2009).
Hu, X. et al. A high throughput multiplex PCR assay for simultaneous detection of seven aminoglycoside-resistance genes in Enterobacteriaceae. BMC Microbiol. 13 , 58. https://doi.org/10.1186/1471-2180-13-58 (2013).
Grape, M., Motakefi, A., Pavuluri, S. & Kahlmeter, G. Standard and real-time multiplex PCR methods for detection of trimethoprim resistance dfr genes in large collections of bacteria. Clin. Microbiol. Infect. 13 , 1112–1118. https://doi.org/10.1111/j.1469-0691.2007.01807.x (2007).
R Core Team. A language and environment for statistical computing. Foundation for Statistical Computing, Vienna, Austria (2021).
Levesque, C., Piche, L., Larose, C. & Roy, P. H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39 , 185–191. https://doi.org/10.1128/AAC.39.1.185 (1995).
White, D. G. et al. The isolation of antibiotic-resistant salmonella from retail ground meats. N. Engl. J. Med. 345 , 1147–1154. https://doi.org/10.1056/NEJMoa010315 (2001).
Download references
The Publication was co-financed within the framework of the Polish Ministry of Science and Higher Education’s program: “Regional Initiative Excellence” in the years 2019–2022 (No. 005/RID/2018/19) “financing amount 12 000 000,00 PLN”.
Author information
Authors and affiliations.
Department of Animal Breeding and Product Quality Assessment. Laboratory of Veterinary Public Health Protection, Poznań University of Life Sciences, Poznań, Poland
Przemysław Racewicz, Michał Majewski, Hanna Biesiada & Sebastian Nowaczewski
Department of Genetics and Animal Breeding, Poznań University of Life Sciences, Poznań, Poland
Marcin Pszczoła & Zofia E. Madeja
Department of Internal Diseases and Veterinary Diagnostics, Poznań University of Life Sciences, Poznań, Poland
Magdalena Kubiak
Lab-Vet Laboratory, Tarnowo Podgórne, Poland
Jarosław Wilczyński & Danuta Wystalska
You can also search for this author in PubMed Google Scholar
Contributions
P.R., M.M. and Z.M. designed experiments and prepared the manuscript. J.W., D.W., M.K., H.B. performed experimental work. P.R. and Z.M. assembled and analyzed the genomic data. M.P. carried out the statistical analyses. S.N. supervised the work progress and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Przemysław Racewicz .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Racewicz, P., Majewski, M., Biesiada, H. et al. Prevalence and characterisation of antimicrobial resistance genes and class 1 and 2 integrons in multiresistant Escherichia coli isolated from poultry production. Sci Rep 12 , 6062 (2022). https://doi.org/10.1038/s41598-022-09996-y
Download citation
Received : 13 December 2021
Accepted : 29 March 2022
Published : 11 April 2022
DOI : https://doi.org/10.1038/s41598-022-09996-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Antibiotic resistance and virulence gene patterns associated with multi drug resistant avian pathogenic escherichia coli (apec) isolated from broiler chickens in india.
- Sandip S. Patel
- Arun C. Patel
- Bhavesh I. Prajapati
Indian Journal of Microbiology (2024)
Global trends in antimicrobial resistance on organic and conventional farms
- Eldon O. Ager
- Tamilie Carvalho
- Jessica L. Hite
Scientific Reports (2023)
The pan-genome of the emerging multidrug-resistant pathogen Corynebacterium striatum
- Hendor N. R. Jesus
- Juliana N. Ramos
- Luis G. C. Pacheco
Functional & Integrative Genomics (2023)
Molecular characterization of multi drug resistant Escherichia coli isolates at a tertiary hospital in Abuja, Nigeria
- Nubwa Medugu
- Mabel Kamweli Aworh
- Siddhartha Thakur
Scientific Reports (2022)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Research article
- Open access
- Published: 11 September 2017
Antimicrobial resistance in Africa: a systematic review
- Birkneh Tilahun Tadesse ORCID: orcid.org/0000-0003-4005-8605 1 , 2 , 3 ,
- Elizabeth A. Ashley 4 ,
- Stefano Ongarello 1 ,
- Joshua Havumaki 1 ,
- Miranga Wijegoonewardena 1 ,
- Iveth J. González 1 &
- Sabine Dittrich 1
BMC Infectious Diseases volume 17 , Article number: 616 ( 2017 ) Cite this article
45k Accesses
263 Citations
167 Altmetric
Metrics details
Antimicrobial resistance (AMR) is widely acknowledged as a global problem, yet in many parts of the world its magnitude is still not well understood. This review, using a public health focused approach, aimed to understand and describe the current status of AMR in Africa in relation to common causes of infections and drugs recommended in WHO treatment guidelines.
PubMed, EMBASE and other relevant databases were searched for recent articles (2013–2016) in accordance with the PRISMA guidelines. Article retrieval and screening were done using a structured search string and strict inclusion/exclusion criteria. Median and interquartile ranges of percent resistance were calculated for each antibiotic-bacterium combination.
AMR data was not available for 42.6% of the countries in the African continent. A total of 144 articles were included in the final analysis. 13 Gram negative and 5 Gram positive bacteria were tested against 37 different antibiotics. Penicillin resistance in Streptococcus pneumoniae was reported in 14/144studies (median resistance (MR): 26.7%). Further 18/53 (34.0%) of Haemophilus influenza isolates were resistant to amoxicillin. MR of Escherichia coli to amoxicillin, trimethoprim and gentamicin was 88.1%, 80.7% and 29.8% respectively. Ciprofloxacin resistance in Salmonella Typhi was rare. No documented ceftriaxone resistance in Neisseria gonorrhoeae was reported, while the MR for quinolone was 37.5%. Carbapenem resistance was common in Acinetobacter spp. and Pseudomonas aeruginosa but uncommon in Enterobacteriaceae .
Our review highlights three important findings. First, recent AMR data is not available for more than 40% of the countries. Second, the level of resistance to commonly prescribed antibiotics was significant. Third, the quality of microbiological data is of serious concern. Our findings underline that to conserve our current arsenal of antibiotics it is imperative to address the gaps in AMR diagnostic standardization and reporting and use available information to optimize treatment guidelines.
Peer Review reports
Internationally, there is a growing concern over antimicrobial resistance (AMR) which is currently estimated to account for more than 700,000 deaths per year worldwide [ 1 ]. If no appropriate measures are taken to halt its progress, AMR will cost approximately 10 million lives and about US$100 trillion per year by 2050 [ 1 ]. In contrast to some other health issues, AMR is a problem that concerns every country irrespective of its level of income and development as resistant pathogens do not respect borders [ 1 , 2 ].
Despite the threat presented by AMR, the 2014 World Health Organization (WHO) and the recent O’Neill report describe significant gaps in surveillance, standard methodologies and data sharing [ 1 , 2 ]. The 2014 WHO report identified Africa and South East Asia as the regions without established AMR surveillance systems [ 2 ]. This lack of quality data is problematic often leading to treatment guidelines that are not adequate for the local situation. The gap in public health capacity is also an issue given the changing resistance mechanisms and the emergence of multidrug-resistant bacteria that can only be detected through systematic screening in quality assured microbiology laboratories [ 3 , 4 ].
One factor contributing to AMR is misuse of antibiotics. Improved malaria diagnostics and the recognition that malaria transmission is decreasing globally has highlighted the lack of tests for other infections and many patients who test negative for malaria are treated with antibiotics indiscriminately [ 5 , 6 , 7 ]. Clinical treatment algorithms like the Integrated Management of Neonatal and Childhood Illnesses (IMNCI) and Integrated Management of Adolescent and Adulthood Illnesses (IMAI) guidelines implemented by the WHO have tried to optimize antibiotic prescription in resource-limited settings, however overuse of antibiotics is still happening [ 8 , 9 , 10 , 11 ]. Following these guidelines amoxicillin or sulfamethoxazole/trimethoprim are the first line drugs for urinary tract infections (UTI) or acute respiratory tract infections. A combination of ampicillin and gentamicin or ceftriaxone are the drugs of choice for treating blood stream infections (BSI) and sulfamethoxazole/trimethoprim or ciprofloxacin are recommended for the treatment of dysentery [ 8 , 9 , 12 ].
A number of recent reviews summarized AMR data in Africa, most recently Leopold et al. (2014) which focused on Sub-Saharan Africa. The authors found a high level of resistance to the commonly used antibiotics in the sub-Saharan African region. For example, 90% of Gram negatives were resistant to chloramphenicol, a commonly used antibiotic. In contrast, resistance to third-generation cephalosporins (like ceftriaxone) was less common, recommending this group for use [ 13 ].
To design suitable local and global interventions, it is important to understand the current status of AMR and identify knowledge gaps. The purpose of this review is to summarize the available information about the occurrence of AMR on the entire African continent and describe laboratory methods currently in use, to identify knowledge gaps and highlight diagnostic needs.
Search strategy
PubMed, EMBASE, Science Daily, the Cochrane Database for Systematic Reviews, African Journals Online Library and Free-text Web Searches using Google Scholar were searched for articles published in English from January 1, 2013 through January 31, 2016. Literature before January 2013 was covered in previous reviews [ 13 , 14 ]. Reference lists of relevant articles were checked for additional titles for inclusion in the review. Key words used for the search were “ Antimicrobial Resistance ”, “ Antimicrobial Susceptibility ”, “ Surveillance ”, “ Diagnostic ”, “ Africa ” and specific names of all African countries. The detailed search strategy, as well as details of the article quality assessment can be found in Additional file 1 .
Selection criteria
Articles reporting AMR prevalence, availability of AMR surveillance systems or diagnostic needs of antibiotic resistance in the whole African region were included. Based on the abstract, articles of all types with any data on etiology and antibiotic susceptibility pattern were included for further screening. Studies were included or excluded following predefined criteria.
Reports on AMR in humans from the African region
Abstracts and full text available in English
Drug sensitivity testing done in a laboratory setting with defined cutoffs for drug susceptibility testing
The denominator as total isolates clearly described for population based studies
Case reports and case series
Reports published before 2013
Studies only focused on malaria, HIV or tuberculosis without AMR information
Studies without information on total studied isolates
Selection procedure
Titles and abstracts of all the articles retrieved through the search were screened. In the event of uncertainty as to whether articles met the criteria for study inclusion they were discussed with two co- authors. Articles selected for full text review were obtained using PubMed, WHO GIFT access, HINARI, institutional websites or by contacting the authors directly. Names of authors from articles in the search results were not blinded for abstract or full-text review.
Data extraction
Data extraction was done using a predesigned and pretested database, developed for the purposes of this review using Microsoft Excel 2013. Information extracted included article information (PMID, first author, year of publication, duration of data collection and country), study design (sample size, age group, hospital acquired or community acquired, number of specimens collected, and clinical syndrome), pathogen identification and antimicrobial susceptibility testing methodology, laboratory accreditation information and antibacterial resistance data.
Article quality assessment
The quality of each article was assessed using a tool modified for the purposes of this study from criteria published by Omulo et al. and the Cochrane guidelines for assessing bias in observational studies. Since a limited number of articles was available, results of the quality assessment were not used for inclusion/exclusion. The quality criteria included 26 items to assess the design, details of sample collection, processing and storage, reporting on AMR methodologies and quality assurance strategies.
Data analysis
We calculated prevalence, median resistance (MR) and inter quartile range (IQR) of resistance for each bacterium-antibiotic combination to calculate a standardized measure from the collected data. Pediatric age was considered less than 18 years and neonatal age less than 28 days. Meta-analysis was not conducted because of the large variability in AMR methodology, geography and the small number of articles available per country. Since the number of studies from hospital/in-patient settings was small, they were combined and median percentages with interquartile ranges were generated. Statistical analyses and visualization were performed using Microsoft Excel 2013, STATA v14 (STATA, College Station, TX, USA) and R-software 3.3.1.
Data and study characteristics
In total, 1704 articles were identified. Of those, 144 studies met the inclusion criteria and were included in the final analysis (Fig. 1 ). Samples from a total of 149,733 patients were analyzed in the selected studies. The majority of the studies were from East Africa (59/144, 40.9%) while the smallest number of studies were from the South African region (6/144, 4.2%) (Fig. 2 ). No suitable report was identified from 23/54, 42.6% countries. While the articles were published between 2013 and 2016, the reported data were collected from 1995 to 2015 with the majority from before 2013 (98/144, 68.1%). Most of the studies (92/144, 63.9%) were cross sectional studies or case series (Table 1 ). Similar numbers of studies were published with susceptibility data for isolates from blood culture (25/144, 17.4%), urine culture (25/144, 17.4%), wound discharge/pus isolates (22/144, 15.3%); and multiple sample types (21/144, 14.6%). More than 80% of the studies fulfilled more than half of the quality parameters (121/144, 84.0%) used to score the articles (Additional file 1 ). Among the different studies, four different methods for susceptibility testing and five different interpretation guidelines were used (Table 1 ).
PRISMA Diagram of the article selection procedure for articles published between January 2013 and January 2016.The review has been registered to the PROSPERO database of systematic reviews on March 1, 2016 ( http://www.crd.york.ac.uk/PROSPERO/myprospero.php ) with ID CRD42016035923
Geographical distribution and number of selected studies between January 2013 and January 2016 in the different African countries. Countries were grouped based on the United Nations Statistics Division classification into Eastern Africa, Southern Africa, Central Africa, Northern Africa and Western Africa
Microbial resistance patterns
The most commonly reported bacterium was Escherichia coli ( 87/144, 60.4%) with the most frequent susceptibility data for gentamicin (77/144, 53.5%), ciprofloxacin (71/144, 49.3%) and sulfamethoxazole/trimethoprim (68/144, 47.2%) (Table 2 ). In total, 13 Gram negative opportunistic pathogens were tested against 37 antibiotics (Table 3 , Additional file 2 ). Overall resistance to commonly used drugs [ 8 , 9 , 12 ], like amoxicillin (MR 72.9%, IQR9.1%–87.3%) and trimethoprim/sulfamethoxazole (MR 75.0%, IQR 49.5%–92.3%) was high. Low to moderate resistance was found to gentamicin (MR 22.1%, IQR 2.0%–45.0%), ciprofloxacin (MR 16.7%, IQR 0%–38.5%) and ceftriaxone (MR 17.2%, IQR 0%-45.0%). Gentamicin resistance in Klebsiella spp., which is naturally resistant to ampicillin, was reported in 1031/2715, 38.0% of tested isolates. Resistance to either ceftriaxone or cefotaxime, which is suggestive of extended–spectrum beta lactamase (ESBL) production was reported in 593/2963, 20.0% and 1051/5395, 19.5% of E. coli as well as 545/1594, 34.2% and 560/1199, 46.7% of the K. pneumoniae isolates, respectively. Imipenem, a drug rarely available in rural Africa, was investigated in 21/144 (14.6%) of the studies and showed the lowest overall MR (MR 3.0%, IQR 0%- 26.6%) (Table 3 ). Of the Acinetobacter isolates reported in three studies, 32.3% (10/31) were resistant to meropenem. Ciprofloxacin resistance in S. Typhi was reported rarely (MR 0%, IQR 0%–11.7%) while ampicillin resistance in Haemophilus influenza was high (MR 100%, IQR76.6%–100%). There was no documented ceftriaxone resistance in N. gonorrhoeae but ciprofloxacin resistance was reported in 37.5% (IQR 30.7%–83.9%) of the isolates.
In the Gram positive group, Coagulase Negative Staphylococcus spp . (CoNS), S. aureus , Streptococcus pneumoniae and Group A streptococcus were the most commonly investigated bacteria (Table 4 ). Methicillin resistant S. aureus (MRSA) was reported in 7/79 (8.9%) of the studies, however as cefoxitin is typically used to screen for MRSA, the MRSA rate is likely underestimated [ 15 ]. Erythromycin resistance in S. aureus was found in 33.9% (IQR 13%–46.4%). Methods used to assess penicillin susceptibility in S. pneumoniae varied: median oxacillin resistance was reported in 40.7% (IQR 0%–55.7%) compared to 26.7% (IQR 8.4%–33.6%) for penicillin and 22.5% (IQR 20%–32.5%) for amoxicillin.Vancomycin showed the lowest resistance pattern for all the tested Gram positive bacteria (Table 4 ).
In the reviewed literature , E. coli was commonly isolated from patients with BSI (17/87, 19.5%), UTI (17/87, 19.5%) and wound infection (16/87, 18.4%). Citrobacter spp. were reported most commonly among patients with UTIs (7/19; 36.8%), followed by BSI (5/19, 26.3%). Two (2/19, 10.6%) [ 16 , 17 ] of the studies which reported on Citrobacter spp. were from HAI; five (5/19, 26.3%) from community acquired infections of which three (3/5) came from pregnant women at antenatal care follow up [ 18 , 19 , 20 ] and two (2/5) had unknown risk factors [ 21 , 22 ]. S . Typhi was most frequently reported from patients with BSI (13/28, 46.4%) followed by patients with diarrhea (6/28, 21.4%). It was also more commonly reported in children below 18 years (12/28, 48.9%) than in adults (2/28, 7.1%) (Table 2 ).
In the current review, the susceptibility results were taken at face value, however there were inconsistencies casting the credibility of several results in doubt. For example a study reporting more than 80% of Proteus isolates ( n = 6) as resistant to imipenem also reported that 80% of them were susceptible to ceftriaxone and 50% were susceptible to vancomycin, a combination that is highly unusual [ 23 ]. There were other unverified reports of highly unusual resistance patterns from some centers, such as penicillin resistant S. pyogenes [ 24 , 25 ] and vancomycin-resistant S. aureus [ 26 , 27 ].
Regional antibiotic resistance patterns
Generally, a lower level of resistance of S. aureus, Klebsiella spp. , E. coli and S. pneumoniae to carbapenems and fluoroquinolones was observed in all the regions as compared to the other antibiotic-bacterium combinations. However, Klebsiella spp. resistance to ciprofloxacin in West Africa was observed to be higher than in other regions. Resistance to the trimethoprim (MR: 33.9%–100%), ampicillin (MR: 7.9%–100%) and penicillin (MR: 0%–75%) was generally high in all regions (Fig. 3 ).
Resistance of selected pathogens to commonly prescribed antibiotics in different regions of Africa. The boxplots in the figure represent the median and interquartile range of resistance reported if at least three studies reported on the combination. Resistance to amoxicillin-clavulanic acid (AMX-CLA), ampicillin, amoxicillin, penicillin, oxacillin, trimethoprim-sulfamethoxazole (TMP-SXT), gentamicin, ceftriaxone, cefoxitin, ciprofloxacin, erythromycin, tetracycline, vancomycin and imipenem were plotted. Antibiotics with no data points in the specific regions are omitted from the plots.Resistance patterns reported using broth dilution minimum inhibitory concentration (MIC), E-test® or VITEK® were included if prevalence could be calculated and were combined with resistance data reported using disk diffusion as this was the main method used. Intermediate susceptible strains were categorized as resistant to simplify the analysis. [ 13 ]MR estimates were not calculated if only one or two studies reported on the specific bacterium-antibiotic combination. a : Resistance of Klebsiella spp. to commonly prescribed antibiotics in different regions of Africa. b : Resistance of S. pneumoniae to commonly prescribed antibiotics in different regions of Africa. c : Resistance of S. aureus to commonly prescribed antibiotics in different regions of Africa. d : Resistance of E. coli to commonly prescribed antibiotics in different regions of Africa
The rise and spread of AMR threatens the effective control and treatment of various bacterial diseases worldwide [ 1 , 2 ]. The achievements gained in reducing mortality and morbidity through early use of antibiotics based on empiric guidelines are in serious jeopardy if appropriate actions are not taken to control AMR [ 28 , 29 ]. Availability of routine and research data on pathogen susceptibilities is an important step towards designing targeted strategies to tackle the global AMR crisis. The current review describes recently (2013–2016) published data on antibiotic drug susceptibility from Africa.
The lack of consistency in the measurement and reporting of susceptibility data makes it difficult to compare findings among different countries and laboratories, sometimes even within one country [ 30 , 31 ]. To address this issue, high income countries have implemented harmonization efforts. For example, laboratories in Europe are encouraged to use the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standard over the Clinical and Laboratory Standards Institute (CSLI) guidelines [ 32 ]. Furthermore, in an effort to enable coherent data synthesis and reporting in January 2016 the British Society for Antimicrobial Chemotherapy (BSAC) actively promoted the EUCAST methods in favour of the current BSAC methodology [ 33 ]. Given the findings of our review, similar harmonization efforts are urgently needed in Africa. Standardizing AMR methods and interpretation guidelines could allow for better comparability of results and improved resistance tracking. Furthermore, improved access to reference laboratories and EQA schemes are needed augmenting the current WHO initiative to scale up the global antimicrobial surveillance system (GLASS) based on country specific priority pathogens [ 34 ]. Currently, in the absence of a uniform laboratory methodology the GLASS goals will be very difficult to meet.
Comparing our findings with previous reviews in the region like Leopold et al. (2014), overall, we identify a similarly high report of resistance to commonly used antibiotics [ 13 ]. The same review reported a high level of resistance of Enterobacteriaceae to ampicillin and co-trimoxazole which is in agreement with our findings. Similarly, resistance to co-trimoxazole and tetracycline by S. pneumoniae was reported to be high. However, discrepancies were observed in various antibiotic/bacterium combinations. For instance, our finding of resistance to chloramphenicol in Salmonella Typhi isolates is lower than that previously reported [ 13 ]. Resistance to oxacillin by S. aureus was also much higher in the current review than the review by Leopold et al. [ 13 ]. The observed differences between data published in 2014 and the current work could indicate a rising pattern in AMR in certain pathogens. However it could also be because of the differences in AMR testing methodologies underlining the need for harmonization of laboratory methods in the region.
A notable finding of this review was the high resistance rate of common causes of UTI to common first line regimes like amoxicillin and sulfamethoxazole/trimethoprim. In the presence of a failing treatment, patients with UTIs are at increased risk of developing renal damage and future risks of renal insufficiency or hypertension [ 35 ]. Similarly, given the resistance profiles in the current review, neonatal sepsis or BSI caused by E. coli , K. pneumoniae and S. aureus are not being effectively treated by first line drugs like ampicillin, aminoglycosides and cephalosporins, which will result in increased mortality in patients with life-threatening infections. The high levels of resistance to amoxicillin and penicillin in S. pneumoniae and H. influenzae are also concerning given that pneumonia is a leading cause of death in children [ 36 , 37 ]. Reported MRSA rates were variable and doubts remain about reliability of identification at all sites which are confirmed by the findings of one Kenyan study which found that MRSA rates dropped dramatically after switching to an automated identification method [ 38 ].
Compared to reports from Asia, quinolone resistance in S . Typhi was rare and, reassuringly, there were no reports of ceftriaxone resistance in N. gonorrhoeae [ 39 ]. Less commonly prescribed antibiotics like imipenem and vancomycin also showed low level resistance and they should be preserved as alternative drugs in severe infections. Most of the imipenem resistance was described in isolates of P. aeruginosa which has been reported from other centers [ 33 ]. Oxacillin resistance should predict penicillin resistance in S. pneumoniae reliably [ 40 , 41 ], however in the current review resistance to oxacillin was much higher than to penicillin, possibly because of the differences in the number of isolates tested for both antibiotics and the use of different cut-offs for meningitis and non-meningitis strains .
The results also yielded data on the susceptibility of less commonly described bacteria like Acinetobacter spp. and Citrobacter spp. Acinetobacter spp. are especially important given their importance in clinical infections and the reported rising trend of resistance, further they have been included in the priority pathogens for global surveillance based on the GLASS initiative [ 42 ]. Citrobacter spp. were reported from hospital and community settings, including in pregnant mothers and can cause UTI which puts pregnant women at risk of preterm labor. Our results would suggest that current frontline treatments are ineffective against most common uro-pathogens.
The limitations of the current review include the exclusion of non-English language reports, as articles from French speaking African countries might have been missed, biasing this review. The representativeness of the data is hard to assess as it is possible that the absence of resistance is not routinely reported and focus is given to reports of resistance. There were very few reports from South Africa, which has a better functioning health system than neighboring countries and better national AMR surveillance. These data were not accessible by our search and therefore larger AMR trends might have been missed. A further limitation is combining AMR results from different patient groups across different countries to compare the data. This approach might have leveled out peaks of resistance in different settings. However, given the observed trends, we believe that the resolution of the obtained data was sufficient to show general developments. Moreover, since case reports were not excluded with intention of capturing as much data as possible in the current review, the findings from the three case (3/144, 2.1% of the studies) might bias the resistance testing results as case reports tend to report on specific multi-drug resistant pathogens. Finally, resistance data obtained with different laboratory methodologies were combined for the purposes of this review. However, as the majority of studies used the disk diffusion method and CLSI guidelines, the impact of the variation in AMR methodology on the validity of the final results is thought to be minimal.
In summary, our review highlights three important findings: first, more than a third of the countries on the continent did not have recent AMR data published in the public domain and only a few of those were surveillance data. Second, a high level of drug resistance exists to commonly prescribed antibiotics in the African continent. Third, the standardization and quality of the microbiological identification and susceptibility testing methods needs to be improved to allow national and international organizations to monitor the extent of the AMR problem. All of the identified areas of concern need urgent attention by the global health community in order to halt the public health threat associated with spreading AMR.
O’Neill J: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations May 2016( http://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf ).
WHO: Antimicrobial Resistance. In: Global Report on surveillance . Edited by WHO. Geneva, Switzerland; 2014.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–8.
Article PubMed CAS Google Scholar
Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia Coli, Belgium , June 2016. Euro Surveill. 2016;21(27)
WHO: World Malaria Report. In . Edited by WHO, vol. http://apps.who.int/iris/bitstream/10665/200018/1/9789241565158_eng.pdf?ua=1 . Switzerland, Geneva; 2015.
Harbarth S, Samore MH. Antimicrobial resistance determinants and future control. Emerg Infect Dis. 2005;11(6):794–801.
Article PubMed PubMed Central Google Scholar
Do NT, Ta NT, Tran NT, Than HM, Vu BT, Hoang LB, van Doorn HR, Vu DT, Cals JW, Chandna A, et al. Point-of-care C-reactive protein testing to reduce inappropriate use of antibiotics for non-severe acute respiratory infections in Vietnamese primary health care: a randomised controlled trial. Lancet Glob Health. 2016;
WHO: Integrated Managment of Neonatal and Childhood Illnesses (IMNCI). Switzerland, Geneva 2014, http://apps.who.int/iris/bitstream/10665/104772/16/9789241506823_Chartbook_eng.pdf?ua=1 .
WHO: Integrated Managment of Adolescent and Adulthood Illnesses (IMAI). Switzerland, Geneva 2009, http://www.who.int/hiv/pub/imai/acute_care.pdf?ua=1 .
Senn N, Rarau P, Salib M, Manong D, Siba P, Rogerson S, Mueller I, Genton B. Use of antibiotics within the IMCI guidelines in outpatient settings in Papua new Guinean children: an observational and effectiveness study. PLoS One. 2014;9(3):e90990.
Article PubMed PubMed Central CAS Google Scholar
Gwimile JJ, Shekalaghe SA, Kapanda GN, Kisanga ER. Antibiotic prescribing practice in management of cough and/or diarrhoea in Moshi municipality, Northern Tanzania: cross-sectional descriptive study. Pan Afr Med J. 2012;12:103.
PubMed PubMed Central Google Scholar
WHO: Pocket book of hospital care for children: Second edition:Guidelines for the management of common childhood illnesses. Switzerland, Geneva 2013, http://www.who.int/maternal_child_adolescent/documents/child_hospital_care/en/ .
Leopold SJ, van Leth F, Tarekegn H, Schultsz C. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan Africa: a systematic review. J Antimicrob Chemother. 2014;69(9):2337–53.
Article CAS PubMed Google Scholar
Le Doare K, Bielicki J, Heath PT, Sharland M. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited countries. J Pediatric Infect Dis Soc. 2015;4(1):11–20.
Article PubMed Google Scholar
Fernandes CJ, Fernandes LA, Collignon P. Cefoxitin resistance as a surrogate marker for the detection of methicillin-resistant Staphylococcus Aureus. J Antimicrob Chemother. 2005;55(4):506–10.
Mengesha RE, Kasa BG-S, Saravanan M, Berhe DF, Wasihun AG. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder teaching and referral hospital, Mekelle Ethiopia. BMC research notes. 2014;7:575.
Mohammed A, Adeshina GO, Ibrahim YK. Incidence and antibiotic susceptibility pattern of bacterial isolates from wound infections in a tertiary hospital in Nigeria. Trop J Pharm Res. 2013;12:617–21.
Google Scholar
Tadesse E, Teshome M, Merid Y, Kibret B, Shimelis T. Asymptomatic urinary tract infection among pregnant women attending the antenatal clinic of Hawassa referral hospital Southern Ethiopia. BMC research notes. 2014;7:155.
Onoh RC, Umeora OUJ, Egwuatu VE, Ezeonu PO, Onoh TJP. Antibiotic sensitivity pattern of uropathogens from pregnant women with urinary tract infection in Abakaliki, Nigeria. Infect Drug Resist. 2013;6:225–33.
Oladeinde BH, Omoregie R, Oladeinde OB. Asymptomatic urinary tract infection among pregnant women receiving ante-natal care in a traditional birth home in Benin City Nigeria. Ethiop J Health Sci. 2015;25(1):3–8.
Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, Muthupandian S. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle hospital Northern Ethiopia. SpringerPlus. 2015;4:314.
Abera B, Kibret M. Azithromycin, fluoroquinolone and chloramphenicol resistance of non-chlamydia conjunctival bacteria in rural community of Ethiopia. Indian J Ophthalmol. 2014;62:236–9.
Eida M, Nasser M, El-Maraghy N, Azab K. Pattern of hospital-acquired pneumonia in intensive care unit of Suez Canal university hospital Egyptian. Journal of Chest Diseases and Tuberculosis. 2015;64:625–31.
Article Google Scholar
Muluye D, Wondimeneh Y, Moges F, Nega T, Ferede G. Types and drug susceptibility patterns of bacterial isolates from eye discharge samples at Gondar University Hospital, Northwest Ethiopia. BMC research notes. 2014;7:292.
Wasihun AG, Zemene Y. Bacterial profile and antimicrobial susceptibility patterns of otitis media in Ayder Teaching and Referral Hospital, Mekelle University, Northern Ethiopia. SpringerPlus. 2015;4:701.
Mengistu A, Gaeseb J, Uaaka G, Ndjavera C, Kambyambya K, Indongo L, Kalemeera F, Ntege C, Mabirizi D, Joshi MP, et al. Antimicrobial sensitivity patterns of cerebrospinal fluid (CSF) isolates in Namibia: implications for empirical antibiotic treatment of meningitis. Journal of Pharmaceutical Policy and Practice. 2013;6
Bayingana C. Antibiotic activity assessment of bacterial strains isolated from urine samples at Butare University Teaching Hospital (Buth) Laboratory. Int J Infect Dis. 2014;21:83.
Gera T, Shah D, Garner P, Richardson M, Sachdev HS. Integrated management of childhood illness (IMCI) strategy for children under five. Cochrane Database Syst Rev. 2016;6:CD010123.
Bhandari N, Mazumder S, Taneja S, Sommerfelt H, Strand TA. Effect of implementation of integrated Management of Neonatal and Childhood Illness (IMNCI) programme on neonatal and infant mortality: cluster randomised controlled trial. BMJ. 2012;344:e1634.
Perdigao-Neto LV, Oliveira MS, Rizek CF, Carrilho CM, Costa SF, Levin AS. Susceptibility of multiresistant gram-negative bacteria to fosfomycin and performance of different susceptibility testing methods. Antimicrob Agents Chemother. 2014;58(3):1763–7.
Article CAS PubMed PubMed Central Google Scholar
Lee M, Chung H-S. Different antimicrobial susceptibility testing methods to detect ertapenem resistance in enterobacteriaceae: VITEK2, MicroScan, Etest, disk diffusion, and broth microdilution. J Microbiol Methods. 2015;112:87–91.
ECDC. Surviellance report: annual report of the European antimicrobial resistance surveillance network (EARS-net). Sweden: Stockholm; 2014. http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-europe-2014.pdf
BSAC: Legacy BSAC Susceptibility Testing Method – no longer supported, for information only. http://www.bsacorguk/stewardship-surveillance/susceptibility/methodologylatestversion/ .
WHO: Antimicrobial Resistance: Global Report on Surveillance. http://www.appswhoint/iris/bitstream/10665/188783/1/9789241549400_engpdf?ua=1 2015.
Park YS. Renal scar formation after urinary tract infection in children. Korean J Pediatr. 2012;55(10):367–70.
Woldu B, Bloomfield GS. Rheumatic heart disease in the twenty-first century. Curr Cardiol Rep. 2016;18(10):96.
Picot VS, Benet T, Messaoudi M, Telles JN, Chou M, Eap T, Wang J, Shen K, Pape JW, Rouzier V, et al. Multicenter case-control study protocol of pneumonia etiology in children: global approach to biological research, infectious diseases and epidemics in low-income countries (GABRIEL network). BMC Infect Dis. 2014;14:635.
Omuse G, Kabera B, Revathi G. Low prevalence of methicillin resistant Staphylococcus Aureus as determined by an automated identification system in two private hospitals in Nairobi Kenya: a cross sectional study. BMC Infect Dis . 2014;14:669.
Dalhoff A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip Perspect Infect Dis. 2012;2012:976273.
Jette LP, Sinave C. Use of an oxacillin disk screening test for detection of penicillin- and ceftriaxone-resistant pneumococci. J Clin Microbiol. 1999;37(4):1178–81.
CAS PubMed PubMed Central Google Scholar
Gertrudis H, María LM, Liliana B, Sigri R, Luz C, Erik M, María EC, Rito Z, Eduardo C, Roger H, et al. Oxacillin disk diffusion testing for the prediction of penicillin resistance in Streptococcus Pneumoniae. Pan Am J Public Health. 2016;40(1):57–63.
Clark NM, Zhanel GG, Lynch JP 3rd. Emergence of antimicrobial resistance among Acinetobacter species: a global threat. Curr Opin Crit Care. 2016;22(5):491–9.
Aduda DSO, Macharia IM, Mugwe P, Oburra H, Farragher B, Brabin B, Mackenzie I. Bacteriology of chronic suppurative otitis media (CSOM) in children in Garissa district, Kenya: a point prevalence study. Int J Pediatr Otorhinolaryngol. 2013;77:1107–11.
Agmy G, Mohamed S, Gad Y, Farghally E, Mohammedin H, Rashed H. Bacterial profile, antibiotic sensitivity and resistance of lower respiratory tract infections in upper Egypt. Mediterranean Journal of Hematology and Infectious Diseases. 2013;5:1–7.
Ahmed EF, Gad GFM, Abdalla AM, Hasaneen AM, Abdelwahab SF. Prevalence of methicillin resistant Staphylococcus Aureus among Egyptian patients after surgical interventions. Surg Infect. 2014;15:404–11.
Ahmed MI, Alsammani MA, Babiker RA. Microbial profile in women with puerperal sepsis in Gadarif state, eastern Sudan. Annals of Tropical Medicine and Public Health. 2013;6:460–4.
Ahmed SF, Klena J, Husain T, Monestersky J, Naguib A, Wasfy MO. Genetic characterization of antimicrobial resistance of Shigella flexneri 1c isolates from patients in Egypt and Pakistan. Ann Clin Microbiol Antimicrob. 2013;12:9.
Akingbade OA, Olasunkanmi OI, Akinjinmi AA, Okerentugba PO, Onajobi BI, Okonko IO. Prevalence and antibiotic profile of Enterobacter species isolated from children with diarrhea in Abeokuta, Ogun state, Nigeria. Stem Cell. 2013;4:1–4.
Alabi AS, Frielinghaus L, Kaba H, Kosters K, Huson MAM, Kahl BC, Peters G, Grobusch MP, Issifou S, Kremsner PG, et al. Retrospective analysis of antimicrobial resistance and bacterial spectrum of infection in Gabon Central Africa. International Journal of Medical Microbiology. 2013;13:455.
Anagaw B, Gezachew M, Biadgelgene F, Anagaw B, Geleshe T, Taddese B, Getie B, Endris M, Mulu A, Unakal C. Antimicrobial susceptibility patterns of Streptococcus Pneumoniae over 6 years at Gondar University hospital, Northwest Ethiopia. Asian Pacific journal of tropical biomedicine. 2013;3:536–41.
Assefa A, Gelaw B, Shiferaw Y, Tigabu Z. Nasopharyngeal carriage and antimicrobial susceptibility pattern of streptococcus pneumoniae among pediatric outpatients at gondar university hospital, north west ethiopia. Pediatrics and Neonatology. 2013;54:315–21.
Babaiwa UF, Onyeagwara NC, Akerele JO. Bacterial tonsillar microbiota and antibiogram in recurrent tonsillitis. Biomedical Research (India). 2013;24:298–302.
Barogui YT, Klis S, Bankole HS, Sopoh GE, Mamo S, Baba-Moussa L, Manson WL, Johnson RC, van der Werf TS, Stienstra Y. Towards rational use of antibiotics for suspected secondary infections in Buruli ulcer patients. PLoS Negl Trop Dis. 2013;7:e2010.
Yeboah-Manu D, Kpeli GS, Ruf M-T, Asan-Ampah K, Quenin-Fosu K, Owusu-Mireku E, Paintsil A, Lamptey I, Anku B, Kwakye-Maclean C, et al. Secondary bacterial infections of buruli ulcer lesions before and after chemotherapy with streptomycin and rifampicin. PLoS Negl Trop Dis. 2013;7:e2191.
van der Meeren BT, Chhaganlal KD, Pfeiffer A, Gomez E, Ferro JJ, Hilbink M, Macome C, van der Vondervoort FJ, Steidel K, Wever PC. Extremely high prevalence of multi-resistance among uropathogens from hospitalised children in Beira, Mozambique. S Afr Med J. 2013;103:382–6.
Musiime V, Cook A, Bakeera-Kitaka S, Vhembo T, Lutakome J, Keishanyu R, Prendergast AJ, Lubwama S, Robertson V, Hughes P, et al. Bacteremia, causative agents and antimicrobial susceptibility among HIV-1-infected children on antiretroviral therapy in Uganda and Zimbabwe. Pediatr Infect Dis J. 2013;32:856–62.
PubMed Google Scholar
Rasamiravaka T, Rasoanandrasana S, Zafindraibe NJJ, Rakoto Alson AO, Rasamindrakotroka A. Evaluation of methicillin-resistant Staphylococcus Aureus nasal carriage in Malagasy patients. J Infect Dev Ctries. 2013;7:318–22.
Kejela T, Bacha K. Prevalence and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus Aureus (MRSA) among primary school children and prisoners in Jimma town Southwest Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:11.
Swierczewski BE, Odundo EA, Koech MC, Ndonye JN, Kirera RK, Odhiambo CP, Cheruiyot EK, Wu MT, Lee JE, Zhang C, et al. Surveillance for enteric pathogens in a case-control study of acute diarrhea in western Kenya. Trans R Soc Trop Med Hyg. 2013;107:83–90.
Sewunet T, Demissie Y, Mihret A, Abebe T. Bacterial profile and antimicrobial susceptibility pattern of isolates among burn patients at Yekatit 12 hospital burn center, Addis Ababa, Ethiopia. Ethiop J Health Sci. 2013;23:209–16.
Seni J, Najjuka CF, Kateete DP, Makobore P, Joloba ML, Kajumbula H, Kapesa A, Bwanga F. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Uganda. BMC research notes. 2013;6:298.
Fahmey SS. Early-onset sepsis in a neonatal intensive care unit in beni suef, Egypt: bacterial isolates and antibiotic resistance pattern. Korean J Pediatr. 2013;56:332–7.
Buhalata SN, Kwesigabo G, Sembuche S, Aboud S, Temu MM, Changalucha JM. Genital tract infections in women attending sexually transmitted infection clinics in Mwanza, north-west Tanzania. Southern African Journal of Epidemiology and Infection. 2013;28:48–54.
Githii S, Revathi G, Muigai A, Kariuki S. Carriage rate and serotypes of streptococcus pneumoniae amongst children in thika hospital, Kenya. African Journal of Laboratory Medicine. 2013;2
Raji MA, Jamal W, Ojemhen O, Rotimi VO. Point-surveillance of antibiotic resistance in Enterobacteriaceae isolates from patients in a Lagos teaching hospital, Nigeria. Journal of Infection and Public Health. 2013;6:431–7.
Omoregie R, Egbe CA, Dirisu J, Ogefere HO. Microbiology of neonatal septicemia in a tertiary hospital in Benin City, Nigeria. Biomarkers and Genomic Medicine. 2013;5:142–6.
Paglietti B, Falchi G, Mason P, Chitsatso O, Nair S, Gwanzura L, Uzzau S, Cappuccinelli P, Wain J, Rubino S. Diversity among human non-typhoidal salmonellae isolates from Zimbabwe. Trans R Soc Trop Med Hyg. 2013;107:487–92.
Page A-L, de Rekeneire N, Sayadi S, Aberrane S, Janssens A-C, Rieux C, Djibo A, Manuguerra J-C, Ducou-le-Pointe H, Grais RF, et al. Infections in children admitted with complicated severe acute malnutrition in Niger. PLoS One. 2013;8:e68699.
Oladeinde BH, Omoregie R, Olley M, Anunibe JA, Onifade AA. A 5 - year surveillance of wound infections at a rural tertiary hospital in Nigeria. Afr Health Sci. 2013;13(2):351–6.
Odongo CO, Anywar DA, Luryamamoi K, Odongo P. Antibiograms from community-acquired uropathogens in Gulu, northern Uganda--a cross-sectional study. BMC Infect Dis. 2013;13:193.
Chiedozie Kingsley O, Onwuezobe Ifeanyi A, Emmanuel Edet A, Chukwuemeka Smart O. Bacteriologic profile and antibiotics susceptibility pattern of suspected septicaemic patients in Uyo, Nigeria. Research Journal of Medical Sciences. 2013;7:35–9.
Esebelahie NO, Newton-Esebelahie FO, Omoregie R. Aerobic bacterial isolates from infected wounds. Afr J Clin Exp Microbiol. 2013;14:155–9.
Mshangila B, Paddy M, Kajumbula H, Ateenyi-Agaba C, Kahwa B, Seni J. External ocular surface bacterial isolates and their antimicrobial susceptibility patterns among pre-operative cataract patients at Mulago National Hospital in Kampala Uganda. BMC ophthalmology . 2013;13:71.
Meyer E, Whitelaw A, Edkins O, Fagan JJ. Chronic otorrhoea: spectrum of microorganisms and antibiotic sensitivity in a south African cohort. South African medical journal = . Suid-Afrikaanse tydskrif vir geneeskunde. 2013;103:471–3.
CAS PubMed Google Scholar
Bissong MEA, Fon PN, Tabe-Besong FO, Akenji TN. Asymptomatic bacteriuria in diabetes mellitus patients in Southwest Cameroon. Afr Health Sci. 2013;13:661–6.
Ibrahim ME, Bilal NE, Magzoub MA, Hamid ME. Prevalence of extended-spectrum β-lactamases-producing Escherichia Coli from hospitals in Khartoum state, Sudan. Oman Medical Journal. 2013;28:116–20.
Lunguya O, Lejon V, Phoba M-F, Bertrand S, Vanhoof R, Glupczynski Y, Verhaegen J, Muyembe-Tamfum J-J, Jacobs JO L et al : Antimicrobial Resistance in Invasive Non-typhoid Salmonella from the Democratic Republic of the Congo: Emergence of Decreased Fluoroquinolone Susceptibility and Extended-spectrum Beta Lactamases PLoS Neglected Tropical Diseases 2013, 7:e2103.
Lengerh A, Moges F, Unakal C, Anagaw B. Prevalence, associated risk factors and antimicrobial susceptibility pattern of Campylobacter species among under five diarrheic children at Gondar University Hospital, Northwest Ethiopia. BMC Pediatr. 2013;13:82.
Iregbu KC, Nwajiobi-Princewill PI. Urinary tract infections in a tertiary hospital in abuja, nigeria. Afr J Clin Exp Microbiol. 2013;14:169–73.
Fadeyibi IO, Raji MA, Ibrahim NA, Ugburo AO, Ademiluyi S. Bacteriology of infected burn wounds in the burn wards of a teaching hospital in Southwest Nigeria. Burns. 2013;39:168–73.
Hancali A, Ndowa F, Bellaji B, Bennani A, Kettani A, Charof R, El Aouad R, A. H, F. N, B. B et al : Antimicrobial resistance monitoring in Neisseria gonorrhoeae and strategic use of funds from the Global Fund to set up a systematic Moroccan gonococcal antimicrobial surveillance programme. Sexually Transmitted Infections 2013, 89 :iv24-iv27.
Hailemariam M, Abebe T, Mihret A, Lambiyo T. Prevalence of Neisseria gonorrhea and their antimicrobial susceptibility patterns among symptomatic women attending gynecology outpatient Department in Hawassa referral hospital, Hawassa, Ethiopia. Ethiop J Health Sci. 2013;23:10–8.
Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:17.
Schaumburg F, Alabi A, Kokou C, Grobusch MP, Köck R, Kaba H, Becker K, Adegnika AA, Kremsner PG, Peters G, et al. High burden of extended-spectrum β-lactamase-producing enterobacteriaceae in Gabon. J Antimicrob Chemother. 2013;68:2140–3.
Donkor ES, Foster-Nyarko E, Enweronu-Laryea CC. Relationship between antibiotic resistance and sickle cell anemia: preliminary evidence from a pediatric carriage study in Ghana. Infection and Drug Resistance. 2013;6:71–7.
Maina EK, Kiiyukia C, Wamae CN, Waiyaki PG, Kariuki S. Characterization of methicillin-resistant Staphylococcus Aureus from skin and soft tissue infections in patients in Nairobi, Kenya. Int J Infect Dis. 2013;17:e115–9.
Chika E, Ifeanyichukwu I, Michael A, Charles E. Susceptibility and detection of extended spectrum β-lactamase enzymes from otitis media pathogens. Am J Infect Dis. 2013;9:24–9.
Dinda V, Gunturu R, Kariuki S, Hakeem A, Raja A, Kimang'a A. Pattern of pathogens and their sensitivity isolated from surgical site infections at the Aga khan university hospital, Nairobi, Kenya. Ethiop J Health Sci. 2013;23:141–9.
Dayie NTKD, Arhin RE, Newman MJ, Dalsgaard A, Bisgaard M, Frimodt-Moller N, Slotved H-C. Penicillin resistance and serotype distribution of Streptococcus Pneumoniae in Ghanaian children less than six years of age. BMC Infect Dis. 2013;13:490.
Dagnew M, Yismaw G, Gizachew M, Gadisa A, Abebe T, Tadesse T, Alemu A, Mathewos B. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC research notes. 2013;6:283.
Coetzee E, Rode H, Kahn D. Pseudomonas Aeruginosa burn wound infection in a dedicated paediatric burns unit. South African journal of surgery Suid-Afrikaanse tydskrif vir chirurgie. 2013;51:50–3.
Christopher A, Mshana SE, Kidenya BR, Hokororo A, Morona D. Bacteremia and resistant gram-negative pathogens among under-fives in Tanzania. Ital J Pediatr. 2013;39:27.
Simango C. Antimicrobial susceptibility of campylobacter species. Southern African Journal of Epidemiology and Infection. 2013;28:139–42.
Bonkoungou IJO, Haukka K, Osterblad M, Hakanen AJ, Traore AS, Barro N, Siitonen A. Bacterial and viral etiology of childhood diarrhea in Ouagadougou Burkina Faso. BMC pediatrics . 2013;13:36.
Acquah SEK, Quaye L, Sagoe K, Ziem JB, Bromberger PI, Amponsem AA. Susceptibility of bacterial etiological agents to commonly-used antimicrobial agents in children with sepsis at the tamale teaching hospital. BMC Infect Dis. 2013;13:89.
Tsai AY, Dueger E, Macalino GE, Montano SM, Mbuchi M, Puplampu N, McClelland RS, Sanchez JL. Neisseria gonorrhoeae (GC) resistance surveillance in selected populations of five countries. Sex Transm Infect. 2013;89
Dau AA, Tloba S, Daw MA: Characterization of wound infections among patients injured during the 2011 Libyan conflict. East Mediterr Health J 2013, 19:356–361.
Adegoke AA, Komolafe AO, Aiyegoro OA. Rare bacterial meningitis among hospitalized children in a teaching hospital. Journal of Pure and Applied Microbiology. 2013;7:973–80.
Negussie A, Mulugeta G, Bedru A, Ali I, Shimeles D, Lema T, Aseffa A. Bacteriological profile and antimicrobial sensitivity pattern of blood culture isolates among septicemia-suspected children at Tikur Anbessa specialized hospital and Yekatit 12 hospital, Addis Ababa, Ethiopia. Crit Care. 2013;17
Shibabaw A, Abebe T, Mihret A. Antimicrobial susceptibility pattern of nasal Staphylococcus Aureus among Dessie referral hospital health care workers, Dessie, Northeast Ethiopia. Int J Infect Dis. 2014;25:e22–5.
Article CAS Google Scholar
Labi A-K, Obeng-Nkrumah N, Addison NO, Donkor ES. Salmonella blood stream infections in a tertiary care setting in Ghana. BMC Infect Dis. 2014;14:3857.
Smith AM, Mthanti MA, Haumann C, Tyalisi N, Boon GPG, Sooka A, Keddy KH. Nosocomial outbreak of salmonella enterica serovar typhimurium primarily affecting a pediatric ward in South Africa in 2012. J Clin Microbiol. 2014;52:627–31.
Abejew AA, Denboba AA, Mekonnen AG. Prevalence and antibiotic resistance pattern of urinary tract bacterial infections in Dessie area North-East Ethiopia. BMC research notes . 2014;7:687.
Aiken AM, Mutuku IM, Sabat AJ, Akkerboom V, Mwangi J, Scott JA, Morpeth SC, Friedrich AW, Grundmann H. Carriage of Staphylococcus Aureus in Thika level 5 hospital Kenya: a cross-sectional study. Antimicrob Resist Infect Control . 2014;3:22.
Akingbade O, Balogun S, Ojo D, Akinduti P, Okerentugba PO, Nwanze JC, Okonko IO, O. A, S. B, D. O et al : Resistant plasmid profile analysis of multidrug resistant Escherichia coli isolated from urinary tract infections in Abeokuta, Nigeria. Afr Health Sci 2014, 14:821–828.
Ako-Nai KA, Ebhodaghe BI, Osho P, Adejuyigbe E, Adeyemi FM, Kassim OO. Preponderance of bacterial isolates in urine of HIV-positive malaria-infected pregnant women with urinary tract infection. Journal of infection in developing countries. 2014;8:1591–600.
Ali MMM, Ahmed SF, Klena JD, Mohamed ZK, Moussa TAA, Ghenghesh KS. Enteroaggregative Escherichia Coli in diarrheic children in Egypt: molecular characterization and antimicrobial susceptibility. Journal of infection in developing countries. 2014;8:589–96.
Conceicao T, Santos Silva I, de Lencastre H, Aires-de-Sousa M: Staphylococcus aureus nasal carriage among patients and health care workers in Sao Tome and Principe. Microbial drug resistance (Larchmont, NY) 2014, 20:57–66.
Cornick JE, Harris SR, Parry CM, Moore MJ, Jassi C, Kamng'ona A, Kulohoma B, Heyderman RS, Bentley SD, Everett DB. Genomic identification of a novel co-trimoxazole resistance genotype and its prevalence amongst Streptococcus Pneumoniae in Malawi. J Antimicrob Chemother. 2014;69:368–74.
Dibua UM, Onyemerela IS, Nweze EI. Frequency, urinalysis and susceptibility profile of pathogens causing urinary tract infections in Enugu state, southeast Nigeria. Rev Inst Med Trop Sao Paulo. 2014;56(1):55–9.
Egyir B, Guardabassi L, Esson J, Nielsen SS, Newman MJ, Addo KK, Larsen AR, B. E, L. G, J. E et al : Insights into nasal carriage of Staphylococcus aureus in an urban and a rural community in Ghana. PLoS One 2014, 9:e96119.
Eleje GU, Adinma JI, Ghasi S, Ikechebelu JI, Igwegbe AO, Okonkwo JE, Okafor CI, Ezeama CO, Ezebialu IU, Ogbuagu CN, et al. Antibiotic susceptibility pattern of genital tract bacteria in pregnant women with preterm premature rupture of membranes in a resource-limited setting. Int J Gynecol Obstet. 2014;127:10–4.
Garedew-Kifelew L, Wondafrash N, Feleke A: Identification of drug-resistant Salmonella from food handlers at the University of Gondar, Ethiopia . BMC research notes 2014, 7:545.
Ibrahim ME, Bilal NE, Hamid ME. Comparison of phenotypic characteristics and antimicrobial resistance patterns of clinical Escherichia Coli collected from two unrelated geographical areas. Global journal of health science. 2014;6:126–35.
Isendahl J, Manjuba C, Rodrigues A, Xu W, Henriques-Normark B, Giske CG, Naucler P. Prevalence of community-acquired bacteraemia in Guinea-Bissau: an observational study. BMC Infect Dis. 2014;14:3859.
Vandepitte J, Hughes P, Matovu G, Bukenya J, Grosskurth H, Lewis DA. High prevalence of ciprofloxacin-resistant gonorrhea among female sex workers in Kampala, Uganda (2008-2009). Sex Transm Dis. 2014;41:233–7.
Moroh J-LA, Fleury Y, Tia H, Bahi C, Lietard C, Coroller L, Edoh V, Coulibaly A, Labia R, Leguerinel I. Diversity and antibiotic resistance of uropathogenic bacteria from Abidjan. Afr J Urol. 2014;20:18–24.
Ramos JM, Perez-Tanoira R, Garcia-Garcia C, Prieto-Perez L, Bellon MC, Mateos F, Tisisano G, Yohannes T, Reyes F, Gorgolas M. Leprosy ulcers in a rural hospital of Ethiopia: pattern of aerobic bacterial isolates and drug sensitivities. Ann Clin Microbiol Antimicrob. 2014;13:47.
Iyamba J-ML, Wambale JM, Lukukula CM, Takaisi-Kikuni NB. High prevalence of methicillin resistant staphylococci strains isolated from surgical site infections in Kinshasa. Pan African Medical Journal. 2014;18
Palle JN, Bassah N, Kamga HLF, Nkwelang G, Akoachere JF, Mbianda E, Nwarie UG, Njunda AL, Assob NJC, Ekane GH, et al. Current antibiotic susceptibility profile of the bacteria associated with surgical wound infections in the buea health district in Cameroon. Afr J Clin Exp Microbiol. 2014;15:27–34.
Kanj SS, Whitelaw A, Dowzicky MJ. In vitro activity of tigecycline and comparators against gram-positive and gram-negative isolates collected from the Middle East and Africa between 2004 and 2011. Int J Antimicrob Agents. 2014;43:170–8.
Kariuki S, Dougan G. Antibacterial resistance in sub-Saharan Africa: an underestimated emergency. Ann N Y Acad Sci. 2014;1323:43–55.
Kihla AJ-FT, Ngunde PJ, Evelyn MS, Gerard N, Ndip RN. Risk factors for wound infection in health care facilities in Buea Cameroon: Aerobic bacterial pathogens and antibiogram of isolates. Pan African Medical Journal . 2014;18:6.
Kouegnigan Rerambiah L, Ndong J-C, Mbakob Mengue Massoua P, Medzegue S, Elisee-Ndam M, Mintsa-Ndong A, Djoba Siawaya JF. Antimicrobial profiles of bacterial clinical isolates from the Gabonese National Laboratory of public health: data from routine activity. Int J Infect Dis. 2014;29:e48–53.
Kibret M, Abera B. Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pacific Journal of Tropical Biomedicine. 2014;4:164–8.
Maltha J, Guiraud I, Kabore B, Lompo P, Ley B, Bottieau E, Van Geet C, Tinto H, Jacobs J. Frequency of severe malaria and invasive bacterial infections among children admitted to a rural hospital in Burkina Faso. PLoS One. 2014;9:e89103.
Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13:14.
Manyahi J, Matee MI, Majigo M, Moyo S, Mshana SE, Lyamuya EF. Predominance of multi-drug resistant bacterial pathogens causing surgical site infections in Muhimbili National Hospital Tanzania. BMC research notes . 2014;7:500.
Moremi N, Mushi MF, Fidelis M, Chalya P, Mirambo M, Mshana SE. Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando medical center. BMC research notes. 2014;7:211.
Mukhtar A, Abdelaal A, Hussein M, Dabous H, Fawzy I, Obayah G, Hasanin A, Adel N, Ghaith D, Bahaa M, et al. Infection complications and pattern of bacterial resistance in living-donor liver transplantation: a multicenter epidemiologic study in Egypt. Transplant Proc. 2014;46:1444–7.
Mulatu G, Beyene G, Zeynudin A. Prevalence of Shigella, salmonella and campylobacter species and their susceptibility patters among under five children with diarrhea in Hawassa town, south Ethiopia. Ethiop J Health Sci. 2014;24:101–8.
Phoba M-F, De Boeck H, Ifeka BB, Dawili J, Lunguya O, Vanhoof R, Muyembe J-J, Van Geet C, Bertrand S, Jacobs J. Epidemic increase in salmonella bloodstream infection in children, Bwamanda, the Democratic Republic of Congo. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2014;33:79–87.
Randrianirina F, Ratsima EH, Ramparany L, Randremanana R, Rakotonirina HC, Andriamanantena T, Rakotomanana F, Rajatonirina S, Richard V, Talarmin A. Antimicrobial resistance of bacterial enteropathogens isolated from stools in Madagascar. BMC Infect Dis. 2014;14:104.
Moyo SJ, Aboud S, Blomberg B, Mkopi N, Kasubi M, Manji K, Lyamuya EF, Maselle SY, Langeland N. High nasal carriage of methicillin-resistant staphylococcus aureus among healthy tanzanian under-5 children. Microb Drug Resist. 2014;20:82–8.
Scherbaum M, Kosters K, Murbeth RE, Ngoa UA, Kremsner PG, Lell B, Alabi A, M. S, K. K, R.E. M et al : Incidence, pathogens and resistance patterns of nosocomial infections at a rural hospital in Gabon. BMC Infect Dis 2014, 14:124.
Swann O, Everett DB, Furyk JS, Harrison EM, Msukwa MT, Heyderman RS, Molyneux EM. Bacterial meningitis in Malawian infants <2 months of age: etiology and susceptibility to World Health Organization first-line antibiotics. Pediatr Infect Dis J. 2014;33:560–5.
Vernet G, Mary C, Altmann DM, Doumbo O, Morpeth S, Bhutta ZA, Klugman KP. Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg Infect Dis. 2014;20(3):434–41.
Wondimeneh Y, Muluye D, Alemu AA, Atinafu A, Yitayew G, Gebrecherkos T, Alemu AA, Damtie D, Ferede G: Urinary tract infection among obstetric fistula patients at Gondar University Hospital, Northwest Ethiopia. BMC Womens Health 2014, 14:12.
Adugna A, Kibret M, Abera B, Nibret E, Adal M. Antibiogram of e. Coli serotypes isolated from children aged under five with acute diarrhea in bahir dar town. Afr Health Sci. 2015;15:656–64.
Abujnah AA, Zorgani A, Sabri MAM, El-Mohammady H, Khalek RA, Ghenghesh KS. Multidrug resistance and extended-spectrum β-lactamases genes among Escherichia Coli from patients with urinary tract infections in northwestern Libya. Libyan Journal of Medicine. 2015;10
Wasihun AG, Wlekidan LN, Gebremariam SA, Welderufael AL, Muthupandian S, Haile TD, Dejene TA. Diagnosis and treatment of typhoid fever and associated prevailing drug resistance in northern Ethiopia. Int J Infect Dis. 2015;35:e96–e102.
Ernest AI, Ng’Walida N, Ndaboine E, Massinde A, Kihunrwa A, Mshana S. Maternal vaginorectal colonization by group B streptococcus and listeria monocytogenes and its risk factors among pregnant women attending tertiary hospital in Mwanza, Tanzania. Tanzania Journal of Health Research. 2015;17
Aamodt H, Mohn SC, Maselle S, Manji KP, Willems R, Jureen R, Langeland N, Blomberg B, H. A, S.C. M et al : Genetic relatedness and risk factor analysis of ampicillin-resistant and high-level gentamicin-resistant enterococci causing bloodstream infections in Tanzanian children. BMC Infect Dis 2015, 15:107.
Abou Shady HM, Bakr AE, Hashad ME, Alzohairy MA. Staphylococcus Aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Braz J Infect Dis. 2015;19(1):68–76.
Alibi S, Ferjani A, Boukadida J. Molecular characterization of extended spectrum beta-lactamases produced by Klebsiella Pneumoniae clinical strains from a Tunisian hospital. Medecine et maladies infectieuses. 2015;45:139–43.
Amissah NA, Glasner C, Ablordey A, Tetteh CS, Kotey NK, Prah I, van der Werf TS, Rossen JW, van Dijl JM, Stienstra Y. Genetic diversity of Staphylococcus Aureus in Buruli ulcer. PLoS Negl Trop Dis. 2015;9:e0003421.
Anago E, Ayi-Fanou L, Akpovi CD, Hounkpe WB, Agassounon-Djikpo Tchibozo M, Bankole HS, Sanni A. Antibiotic resistance and genotype of beta-lactamase producing Escherichia Coli in nosocomial infections in Cotonou Benin. Annals of clinical microbiology and antimicrobials . 2015;14:5.
Ayepola OO, Olasupo NA, Egwari LO, Becker K, Schaumburg F. Molecular characterization and antimicrobial susceptibility of Staphylococcus Aureus isolates from clinical infection and asymptomatic carriers in Southwest Nigeria. PLoS One. 2015;10(9):e0137531.
Kwambana-Adams B, Darboe S, Nabwera H, Foster-Nyarko E, Ikumapayi UN, Secka O, Betts M, Bradbury R, Wegmüller R, Lawal B, et al. Salmonella infections in the Gambia, 2005-2015. Clin Infect Dis. 2015;61:S354–62.
Kraef C, Alabi AS, Peters G, Becker K, Kremsner PG, Rossatanga EG, Mellmann A, Grobusch MP, Zanger P, Schaumburg F. Co-detection of Panton-valentine leukocidin encoding genes and cotrimoxazole resistance in Staphylococcus Aureus in Gabon: implications for HIV-patients' care. Front Microbiol. 2015;6
Langendorf C, Le Hello S, Moumouni A, Gouali M, Mamaty A-A, Grais RF, Weill F-X, Page A-L. Enteric bacterial pathogens in children with diarrhea in niger: diversity and antimicrobial resistance. PLoS One. 2015;10:e0120275.
Mofolorunsho CK, Ocheni M, Omatola CA, Agieni AG. Staphylococcus Aureus prevalence and antibiotic susceptibility profile in anyigba, north-central Nigeria. Am J Infect Dis. 2015;11:93–7.
Shehab El-Din EMR, El-Sokkary MMA, Bassiouny MR, Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: a study from Egypt. Biomed Res Int. 2015;2015
Nwankwo EO, Magaji NS, Tijjani J. Antibiotic susceptibility pattern of extended spectrum betalactamase (ESBL) producers and other bacterial pathogens in Kano, Nigeria. Trop J Pharm Res. 2015;14:1273–8.
Iliyasu G, Habib AG, Aminu MB. Antimicrobial susceptibility pattern of invasive pneumococcal isolates in north West Nigeria. J Global Infect Dis. 2015;7:70–4.
Tesfaw G, Kibru G, Mekonnen D, Abdissa A. Prevalence of group a β-haemolytic streptococcus among children with pharyngitis in Ethiopia. Eur Respir J. 2015;46
Hamdan HZ, Kubbara E, Adam AM, Hassan OS, Suliman SO, Adam I. Urinary tract infections and antimicrobial sensitivity among diabetic patients at Khartoum Sudan. Annals of clinical microbiology and antimicrobials . 2015;14:26.
Mandomando I, Bassat Q, Sigaúque B, Massora S, Quintó L, Ácacio S, Nhampossa T, Vubil D, Garrine M, Macete E, et al. Invasive salmonella infections among children from rural Mozambique, 2001-2014. Clin Infect Dis. 2015;61:S339–45.
Onwuezobe IA, Orok FE. Extended spectrum beta-lactamase producing uropathogens in asymptomatic pregnant women attending antenatal care in an urban community secondary health facility. Afr J Clin Exp Microbiol. 2015;16:49–53.
Iregbu KC, Eze SO. Pseudomonas Aeruginosa infections in a tertiary hospital in Nigeria. Afr J Clin Exp Microbiol. 2015;16:33–6.
Kityamuwesi R, Muwaz L, Kasangaki A, Kajumbula H, Rwenyonyi CM. Characteristics of pyogenic odontogenic infection in patients attending Mulago hospital Uganda: A cross-sectional study. BMC Microbiology . 2015;15:46.
Kalonji LM, Post A, Phoba M-F, Falay D, Ngbonda D, Muyembe J-J, Bertrand S, Ceyssens P-J, Mattheus W, Verhaegen J, et al. Invasive salmonella infections at multiple surveillance sites in the Democratic Republic of the Congo, 2011-2014. Clin Infect Dis. 2015;61:S346–53.
El Bouamri MC, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella Pneumoniae and the emergence of carbapenem-resistant strains: a retrospective study from a university hospital in Morocco, North Africa. Afr J Urol. 2015;21:36–40.
Ntirenganya C, Manzi O, Muvunyi CM, Ogbuagu O. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in Rwanda. Am J Trop Med Hyg. 2015;92:865–70.
Ochieng N, Okechi H, Ferson S, Albright AL. Bacteria causing ventriculoperitoneal shunt infections in a Kenyan population. J Neurosurg Pediatr. 2015;15:150–5.
Onken A, Said AK, Jorstad M, Jenum PA, Blomberg B. Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja Zanzibar. PLoS One . 2015;10:e0145632.
Peterside O, Pondei K, Akinbami FO. Bacteriological profile and antibiotic susceptibility pattern of neonatal sepsis at a teaching hospital in Bayelsa state, Nigeria. Tropical Medicine and Health. 2015;43:183–90.
Preziosi M, Zimba TF, Lee K, Tomas M, Kinlin S, Nhatave-Paiva C, Bene R, Paunde T, Lopes H, Kalkhoff S, et al. A prospective observational study of bacteraemia in adults admitted to an urban Mozambican hospital. S Afr Med J. 2015;105:370–4.
Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4
Saeed A, Abd H, Sandstrom G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J Med Microbiol. 2015;64:432–7.
Okoje-Adesomoju VN, Ifesanya JU, Alonge TO. Antimicrobial susceptibility pattern of oral microbial isolates among pregnant women in Ibadan south-east local government area, Nigeria. Afr J Biomed Res. 2015;18:29–35.
Mulu W, Yimer M, Zenebe Y, Abera B. Common causes of vaginal infections and antibiotic susceptibility of aerobic bacterial isolates in women of reproductive age attending at Felegehiwot referral hospital, Ethiopia: a cross sectional study. BMC Womens Health. 2015;15
Maina D, Omuse G, Revathi G, Adam RD. Spectrum of microbial diseases and resistance patterns at a private teaching Hospital in Kenya: implications for clinical practice. PLoS One. 2016;11:e0147659.
Odetoyin BW, Hofmann J, Aboderin AO, Okeke IN: Diarrhoeagenic Escherichia coli in mother-child Pairs in Ile-Ife, South Western Nigeria. BMC Infect Dis 2016.
Onanuga A, Selekere TL. Virulence and antimicrobial resistance of common urinary bacteria from asymptomatic students of Niger Delta University, Amassoma, Bayelsa state, Nigeria. Journal of Pharmacy and Bioallied Sciences. 2016;8:29–33.
Abdelaziz ZA, Ibrahim ME, Bilal NE, Hamid ME. Vaginal infections among pregnant women at Omdurman maternity Hospital in Khartoum, Sudan. Journal of Infection in Developing Countries. 2014;8:490–7.
Olorunmola FO, Kolawole DO, Lamikanra A. Antibiotic resistance and virulence properties in escherichia coli strains from cases of urinary tract infections. African Journal of Infectious Diseases. 2013;7:1–7.
Pondei K, Fente BG, Oladapo O. Current microbial isolates from wound swabs, their culture and sensitivity pattern at the Niger Delta University teaching hospital, Okolobiri, Nigeria. Tropical Medicine and Health. 2013;41:49–53.
Maina D, Makau P, Nyerere A, Revathi G. Antimicrobial resistance patterns in extended-spectrum β-lactamase producing Escherichia Coli and Klebsiella Pneumoniae isolates in a private tertiary hospital, Kenya. Microbiology Discovery. 2013;1
Sanford Guide - Trusted infectious disease recommendations: http://www.sanfordguide.com/stewardship/ .
Download references

Acknowledgments
The authors would like to acknowledge Dr. Timothy Rodwell for commenting on the systematic review protocol as well as Piedra Lightfoot, Beatrice Gordis, Dr. Bill Rodriguez and Dr. Sophia Georghiou for critical review of the manuscript.
Ethics and consent to participate
Not applicable.
The Australian, UK, Dutch and Swiss Government and the WHO/TDR-program.
Availability of data and materials
Data supporting our findings can be found through the corresponding author (email: [email protected]) or the senior author (email: [email protected]).
Author information
Authors and affiliations.
Foundation for Innovative New Diagnostics (FIND), Campus Biotech Building B2 Level 0, 9 Chemin des Mines, 1202, Geneva, Switzerland
Birkneh Tilahun Tadesse, Stefano Ongarello, Joshua Havumaki, Miranga Wijegoonewardena, Iveth J. González & Sabine Dittrich
College of Medicine and Health Sciences, Department of Pediatrics, Hawassa University, Hawassa, Ethiopia
Birkneh Tilahun Tadesse
Special Programme for Research & Training in Tropical Diseases (TDR), World Health Organization, Avenue Appia 20, 1211, 27, Geneva, Switzerland
Myanmar-Oxford Clinical Research Unit (MOCRU), Yangon, Myanmar
Elizabeth A. Ashley
You can also search for this author in PubMed Google Scholar
Contributions
BTT, IJG and SD conceived the idea, prepared the systematic review protocol and were involved in the literature search, screening and extraction of data. BTT performed the data analysis and developed the first draft of the manuscript. SD, IJG, SO, MW, JH, EA provided critical review of the manuscript and contributed to the final version. All the authors read and approved the final version of the manuscript.
Corresponding author
Correspondence to Birkneh Tilahun Tadesse .
Ethics declarations
Competing interests.
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:.
Detailed Methododologies (DOCX 24 kb)
Additional file 2:
Proportion of resistance of bacterial isolates to tested antibiotics by region and country (DOCX 263 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
Tadesse, B.T., Ashley, E.A., Ongarello, S. et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis 17 , 616 (2017). https://doi.org/10.1186/s12879-017-2713-1
Download citation
Received : 21 February 2017
Accepted : 04 September 2017
Published : 11 September 2017
DOI : https://doi.org/10.1186/s12879-017-2713-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Antimicrobial resistance
- Systematic review
BMC Infectious Diseases
ISSN: 1471-2334
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Click through the PLOS taxonomy to find articles in your field.
For more information about PLOS Subject Areas, click here .
Loading metrics
Open Access
Peer-reviewed
Research Article
Awareness and knowledge of antimicrobial resistance and factors associated with knowledge among adults in Dessie City, Northeast Ethiopia: Community-based cross-sectional study
Roles Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing
* E-mail: [email protected]
Affiliation Department of Social and Administrative Pharmacy, School of Pharmacy, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia
Roles Conceptualization, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing
Affiliation Department of Pharmacy, College of Medicine and Health Sciences, Wollo University, Dessie, Ethiopia
- Wudneh Simegn,
- Getachew Moges

- Published: December 30, 2022
- https://doi.org/10.1371/journal.pone.0279342
- Reader Comments
Antimicrobial resistance is an important global health challenge. The current study aimed to assess the level of awareness and knowledge of antimicrobial resistance and factors associated with knowledge among adults in Dessie City, Ethiopia.
A community-based cross-sectional study was conducted among 407 adults in Dessie City from June to July 2021. A systematic random sampling technique was used to select respondents, and Google Form was used to collect data online. The data was analyzed by SPSS Version 26. The associated factors of knowledge of antimicrobial resistance were identified by using bivariate and multivariable logistic regression. Independent variables with a P-value <0.2 were selected as candidate variables for multivariable logistic regression. Those variables with a P-value <0.05 were declared statistically significant factors.
Out of the required sample sizes, four hundred and seven participants were enrolled, giving a response rate of 99.3%. One hundred and fifty-two (37.3%) respondents were females. Nearly one-third of the respondents (28.3%) have taken antibiotics in the last 6 months. In this study, 73.7% of study participants were aware of the existence of germs; 58.2% were aware of the existence of antibiotic resistance to bacteria; 47.7% were aware of the existence of drug resistance; 39.8% were aware of the existence of antimicrobial resistance; and 36.6% were aware of the existence of antibiotic resistance. Sixty-four (15.7%) respondents were not aware of any of the above terms. Sixty (14.7%) of the respondents were not aware of any risk factor for antimicrobial resistance. About 63 (15.5%) of the respondents did not know the consequences of antimicrobial resistance. Two hundred and thirty-eight (58.5%) respondents had good knowledge of antimicrobial resistance. In this study, being male (AOR = 1.99; 95% CI: 1.23,3.20), college and above educational level (AOR = 3.50; 95% CI: 1.08,11.39), grade 11–12 educational level (AOR = 3.73; 95% CI: 1.20,11.61), getting advice from health professionals about how to take antibiotics (AOR = 1.84; 95% CI:1.07,3.17), using health professionals as a source of information on antibiotics (AOR = 2.51; 95% CI: 1.48,4.25), and taking antibiotics without prescription (AOR = 1.86; 95% CI: 1.04,3.30) were significantly associated with good knowledge of antimicrobial resistance.
The study identified low awareness and knowledge of antimicrobial resistance among adults. Being male, higher educational level, getting advice from health professionals about how to take antibiotics, using health professionals as a source of information on antibiotics, and taking antibiotics without a prescription were significantly associated with good knowledge of antimicrobial resistance. Educational campaigns would be highly desirable for the public to improve their awareness and knowledge of antimicrobial resistance.
Citation: Simegn W, Moges G (2022) Awareness and knowledge of antimicrobial resistance and factors associated with knowledge among adults in Dessie City, Northeast Ethiopia: Community-based cross-sectional study. PLoS ONE 17(12): e0279342. https://doi.org/10.1371/journal.pone.0279342
Editor: Muhammad Shahzad Aslam, Xiamen University - Malaysia Campus: Xiamen University - Malaysia, MALAYSIA
Received: March 28, 2022; Accepted: December 4, 2022; Published: December 30, 2022
Copyright: © 2022 Simegn, Moges. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: All relevant data are within the manuscript and its Supporting Information files.
Funding: The author(s) received no specific funding for this work.
Competing interests: The authors have declared that no competing interests exist.
Abbreviations: AMR, Antimicrobial Resistance; WHO, World Health Organization; CI, Confidence Interval
Introduction
As it is defined by the World Health Organization (WHO), appropriate use of antimicrobials is the cost-effective use of antimicrobials that maximizes clinical therapeutic effect while minimizing both drug-related toxicity and the development of resistance [ 1 , 2 ]. Antimicrobial resistance is becoming the main global health challenge [ 3 – 6 ]. Globally, drug resistance causes an estimated 700,000 deaths each year [ 7 , 8 ]. It is predominantly high in low and middle income countries and is partly related to high levels of antimicrobial use [ 7 , 9 – 14 ]. Besides, the environment is also increasingly affected by the global spread of clinically relevant antimicrobial resistant organisms [ 4 , 15 ], which impacts treatment outcomes [ 16 , 17 ]. The other major factors contributing to the emergency and spread of AMR are misuse and overuse of antimicrobial agents [ 12 , 18 ], high load of infectious diseases, a deprived infection-control policy, poor-quality medicines, inadequate knowledge of AMR, misdiagnosis, and lack of laboratories for antibiotic susceptibility tests [ 7 , 19 – 21 ].
Several educational campaigns have been implemented worldwide to reduce AMR with different levels of effectiveness [ 11 , 19 , 22 , 23 ]. The WHO is organizing a global movement to raise awareness of antibiotic resistance and encourage best practices among the public, health, and agricultural professionals to avoid the further emergence and spread of antibiotic resistance by implementing antimicrobial stewardship [ 9 , 18 , 24 ]. Due to the common prevalence of infectious diseases in Ethiopia [ 24 , 25 ], there is an irrational use of antibiotics by the public, patients, and healthcare providers [ 21 , 26 ]. Despite this, no national antimicrobial control system or policy has been implemented to reduce AMR [ 27 ].
Several studies have been conducted to assess awareness and knowledge of AMR among the public. A cross-sectional survey in the district of Sialkot, Pakistan reported that more than half of the community participants (55.6%) had poor knowledge of AMR [ 28 ]. A cross-sectional study done at Felege Hiwot Hospital, Ethiopia among patients showed that 42.3% had poor knowledge about AMR [ 29 ]. A recent study in Kemissie Town, Northeast Ethiopia, reported that 41.6% of the community had awareness of AMR [ 24 ].
Rational use of antimicrobials is the main strategy to prevent AMR [ 30 ]. To achieve this, measuring the level of awareness and knowledge of AMR among the public will play a great role. This will help to identify the gaps for taking measures to promote appropriate use of antimicrobials and prevent the further development of antimicrobial resistance. It will also be important to plan the implementation of educational campaigns by health sector administrators. As there were no previous local studies that investigated the awareness and knowledge of AMR among adults, the authors assessed the level of awareness and knowledge of AMR and factors associated with knowledge among adults in Dessie City, Ethiopia.
Study area, design, and period
A community-based cross-sectional study was conducted in Dessie City, Northeast Ethiopia. Dessie is a multi-ethnic City located in the South Wollo zone of Amhara Regional State, 401 km away from Addis Ababa, the capital City of Ethiopia. The projection plan commission reported that there would be 48,144 people in 2021. There were 20 urban and 8 rural Kebeles (smallest units of administration in Ethiopia), which were administered in the City. The City had five hospitals (one public referral, one public general, and three private hospitals), eight health centers, and twenty-seven private clinics. In the hospitals, there were AMR stewardship committees and an infection prevention committee. The committees in each hospital provided in-service training to the health professionals and an educational campaign to patients in the morning session about AMR, but there was no well-structured educational program. There is no educational campaign given outside of hospitals to the general public. The study was conducted from June 30 to July 30, 2021.
All residents living in Dessie City were used as the source population. The study population consisted of residents who were present at the time of data collection. Residents who were health professionals, those who were severely ill, those who were unable to speak, and those who had lived less than six months were excluded.
Sample size calculation and sampling technique
The sample size was calculated using a single population proportion formula with the assumptions of prevalence of AMR awareness (41.6%) in the previous study [ 24 ], a 95% confidence interval and a margin of error (d) of 5%. After adding a non-response rate of 10%, the final sample size for this study was 410. We randomly selected 5 Kebeles (Dawudo, Ager Gizat, Robit, Tekuam, and Kelemmeda) from the total of 20 urban Kebeles in the City, and 3 Kebeles (Gerado Tesfanech, Boru Meda, and Gerado Kelina) from the total of 8 Kebeles in the rural parts. A stratified sampling technique was used to allocate the study participants proportionately to the urban and rural settings. The current number of households was taken from the City administration. The list of the households with their respective addresses was taken at each Kebele administrative office. From each stratum, samples were taken proportionally to their size by taking the number of households as the sampling frame. Finally, we used 361 study participants from the urban parts of the City and 46 study participants from the rural parts of the City. The households from each Kebele of an urban stratum and from each Kebele of a rural stratum were selected using a systematic random sampling technique. The sampling interval for each Kebele was determined by dividing the total number of households in each Kebele into its proportionally allocated sample size. Then, every K th value of households was interviewed by randomly selecting the first household. The flow chart of sampling technique is included below ( Fig 1 ).
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.pone.0279342.g001
Data collection instrument and data collection procedure
The data was collected using a semi-structured interview-based questionnaire, which was adapted from the WHO level of antimicrobial resistance public awareness survey and another study [ 18 , 24 ]. The questionnaire included four parts. Part 1 included socio-demographic characteristics such as sex, age, educational level, occupation, and monthly income. Part 2 included items related to antibiotics use. Part 3 included questions used to assess awareness of antimicrobial resistance, including the risk factors and consequences of AMR. Part 4 included eight true/false questions to assess knowledge of AMR. As the data was collected during the peak COVID-19 outbreak, we used an electronic online Google form for data collection to minimize the spread of the pandemic. Two pharmacists were recruited to collect the data. They interviewed the eligible study participants and filled in the electronic online Google form using a smart phone. The data collectors contacted the head of the household for an interview. If the head of the household was not present at the time of data collection, any member of the household who was 18 years of age or older was recruited for the study. If any member of a household was not available during data collection, the next household was selected.
Data quality control
A pretest was done among 20 residents outside of the study area to check the clarity of the questions, and we amended the few terms in the socio-demographic parts. The data collectors were trained about the purpose of the study and ethical issues in the process of data gathering. The principal investigator was actively involved in the data collection process. The questionnaire was prepared in English and translated to the local language (Amharic) and then back-translated into the English language by fluent speakers of the two languages to check the consistency.
Variables of the study
Socio-demographic characteristics such as gender, age, residence, religion, marital status, educational level, occupation, and average monthly income were included. Information related to antibiotic use, such as the last time taking antibiotics, the sources of antibiotics, whether respondents got advice while taking antibiotics, the source of information on antibiotics, whether they suffered from different microbial infections, practice of self-medication with antibiotics, awareness, and knowledge of AMR were included.
Measuring techniques
The measuring techniques for awareness of AMR describe the frequency and percentage score of each question. The measuring technique for knowledge of AMR was based on the World Health Organization (WHO) level of measurement of antimicrobial resistance among communities [ 18 ]. Respondents were provided with true or false answers to the eight questions, which were used to grade the respondents’ knowledge. A score of 1 was assigned for the correct responses and 0 for the incorrect ones. Then, the percentage score was determined. Study participants who scored less than 75% were considered to have poor AMR knowledge, while those who scored 75% and above were considered to have good AMR knowledge. This cut-off point was based on the national survey of public knowledge of AMR conducted in Nigeria [ 31 ].
Terminologies
Antimicrobial resistance..
Antimicrobial resistance occurs when bacteria, viruses, fungi and parasites change over time and no longer respond to medicines making infections harder to treat and increasing the risk of disease spread, severe illness and death [ 32 ].
Antibiotic resistance.
Antibiotic resistance occurs when germs such as bacteria develop the ability to resist drugs that are designed to kill them [ 33 ].
Refers to the microscopic bacteria, viruses, fungi, and protozoa that can cause disease [ 34 ].
Drug resistance.
The ability of disease-causing germs (e.g. bacteria or viruses) to continue multiplying despite the presence of drugs that usually kill them [ 32 ].
Antibiotic-resistant bacteria.
The bacteria that are not killed by antibiotics and can continue to multiply presents a substantial threat to the control of infectious diseases [ 35 ].
Statistical analysis
The data was analyzed using SPSS Version 26. The mean with standard deviation (SD) and frequency with percent were computed to describe the results of the study. Logistic regression was used to explain the association between knowledge of AMR and the independent variables. Independent variables with a p-value < 0.2 were selected as candidate variables for multivariable logistic regression. Variables with a p-value < 0.05 and 95% CI were treated as a significant factors for knowledge of AMR.
Ethical approval and consent to participate
Ethical clearance was obtained from Wollo University, College of Medicine and Health Sciences, department of pharmacy Ethical review board with reference number: WU Phar/266/2013. A letter of cooperation was written to the Dessie City Administration office. Permission letters were obtained from each Kebele administration office. Written consent was obtained from each study participant. Confidentiality of the information collected was maintained by omitting any personal identifier from the data collection tool. This study was conducted according to the declaration of Helsinki.
Socio-demographic characteristics of the study participants
In the current study, 407 study participants were enrolled with a response rate of 99.5% of which 152 (37.3%) were females. The mean age with standard deviation of the participants was 35.63±8.63 years and ranged from 20–68 years. Thirty (7.4%) participants were unable to read and write, while most of the participants attended grades 11–12 (30.2%). Majority (38.8%) of the study participants were merchants, followed by employees (24.3%) and housewives (16.2%) ( Table 1 ).
https://doi.org/10.1371/journal.pone.0279342.t001
Reported antibiotics use by the study participants
Almost one-third (28.3%) of the respondents took antibiotics in the last 6 months. The majority of respondents got antibiotics from retail outlet pharmacies (66.3%). More than half (60.2%) of the respondents reported that they were advised by health professionals regarding antibiotics. The major sources of information about antibiotics were healthcare professionals (64.9%), followed by friends and family (46.2%), and previous experiences (38.3%). The majority (71.5%) of the respondents reported that they stop taking antibiotics when they have taken all the antibiotics as directed, while more than one-third (36.6%) reported that they discontinued their antibiotics when they felt better ( Table 2 ).
https://doi.org/10.1371/journal.pone.0279342.t002
Awareness and knowledge of antimicrobial resistance
Of the total respondents included in this study, 73.7% of the respondents were aware of the existence of germs; 58.2% of the respondents were aware of the existence of antibiotic-resistant bacteria; 47.7% of the respondents were aware of the existence of drug resistance; 39.8% of the respondents were aware of the existence of antimicrobial resistance; and 36.6% of the respondents were aware of the existence of antibiotic resistance, while only 15.7% of the respondents were not aware of any of the existence of the above terms. Most of the respondents (70.8%) were aware that sharing antibiotics with others was a risk factor for AMR. Other risk factors mentioned by the respondents were failure to complete the course of therapy (61.4%), taking antibiotics without a prescription (54.3%), over or under use of antibiotics (43.7%) and taking antibiotics without considering the dose and time gap (40.5%). Sixty (14.7%) of the respondents were not aware of any of the risk factors for AMR. No cure for the diseases (77.1%), need for expensive drugs (63.9%), increased intensity and duration of the diseases (45.9%) and decreased antibiotic activity (45.2%) were the consequences of AMR reported by respondents. About 15.5% of the respondents never knew of any consequences related to AMR. In the current study, the mean knowledge score of all participants was 5.40 (95% CI: 5.28, 5.51). Two hundred and thirty-eight (58.5%) respondents had good knowledge of AMR ( Table 3 ).
https://doi.org/10.1371/journal.pone.0279342.t003
Association of different factors with knowledge of antimicrobial resistance
Gender, age, residence, educational level, the last time to take antibiotics, using health facilities as a source of antibiotics, relying upon health professionals to get advice about how to take antibiotics, using health professionals as a source of information for antibiotics, relying upon previous experience for source of information for antibiotics, taking antibiotics without prescription, and suffering from different microbial infections were candidate variables for multivariable logistic regression (p-value<0.2).
In the final model, male study participants had 1.99-folds (AOR = 1.99; 95% CI: 1.23,3.20) better knowledge of AMR than females. Study participants with grade 11–12 educational level had 3.73 odds (AOR = 3.73; 95% CI: 1.20,11.61) and those with college and above educational level had 3.50 times (AOR = 3.50; 95% CI: 1.08,11.39) better knowledge of AMR than study participants who were unable to read and write. Study participants who were relying upon health professionals to get advice about how to take antibiotics had 1.84 times (AOR = 1.84; 95% CI: 1.07,3.17) better knowledge of AMR than those study participants who did not get advice from health professionals. Study participants who ever used health professionals as a source of information on antibiotics had 2.51 times (AOR = 2.51; 95% CI: 1.48,4.25) and those who used antibiotics without a prescription had 1.86 (AOR = 1.86; 95% CI: 1.04,3.30) better knowledge of AMR than their counterparts ( Table 4 ).
https://doi.org/10.1371/journal.pone.0279342.t004
The current study assessed awareness and knowledge of AMR and factors associated with knowledge among adults in Dessie City, northeast Ethiopia. Nearly three-fourths of the respondents (73.5%) were aware of the term "germ". More than half of the study participants (58.2%) were aware of antibiotic resistant bacteria. Under half of the study participants were aware of antimicrobial resistance (39.8%) and antibiotic resistance (36.6%). The finding indicated a lower level of AMR awareness among the adults in Dessie City. The root causes of the low level of AMR awareness might be due to lack of information about the risk factors and the consequences of AMR, lack of engagement with the wider healthcare workforce, socio-economic factors and lack of an AMR educational campaign for the public [ 36 – 38 ]. Healthcare administrators should have an integrated approach by prioritizing AMR awareness creation and declaring the individual’s accountability could be the possible mechanism to bring changes in the awareness of AMR. To address the problems, increasing public education by using a nationwide television, radio, and social media campaign might be expected. This should target the masses to educate them about AMR, its causes, and effects by using more holistic public enlightenment programs [ 31 , 37 , 39 – 41 ].
Awareness of the term antibiotic resistance was lower than studies in Kemssie (59.4%) [ 24 ], in Nigeria (56.5%) [ 31 ], in Romania (85.14%) [ 42 ], and in China (95%) [ 43 ]. The study done in China was for urban people and those with a higher level of education, but the current study was conducted with respondents having different socio-demographic characteristics. Our study identified about 64 (15.7%) of the respondents were not aware of any term related to AMR. It was almost similar to the national survey in Nigeria (17.0%) [ 31 ].
In the present study, about 288 (70.8%) of the respondents were aware that sharing antibiotics with others is a risk factor for AMR. This is significantly higher than a report in Kemssie (17.7%) [ 24 ]. Even though better awareness of the risk of sharing antibiotics is observed in the current study, an educational campaign among the public should be given to keep the momentum. The educational campaign should address the reasons for taking antibiotics to only specific individuals prescribed for a particular episode of illness [ 37 ]. A higher proportion of the respondents (61.4%) were also aware that failure to complete the course of therapy is a risk for antibiotic resistance, which is higher than the one reported in Kemssie (23.7%) [ 24 ]. The educational campaign should also include the importance of taking the full prescription as prescribed [ 37 ]. Only about 54.3% of study participants were aware that taking antibiotics without a prescription is a risk factor for AMR. The finding showed an urgent need to revisit rules on antibiotic prescriptions and OTC sales of antibiotics. As there is no strict rule to control the selling of antibiotics without a prescription at the community pharmacy in Ethiopia, antibiotics are used without having been prescribed. This problem could be reduced by educational campaigns for the public and setting up and controlling policies and regulations regarding antibiotic sales [ 37 , 39 , 44 ]. In addition, involving all pharmacists to educate all patients every time during dispensing antibiotics would be important. Not getting cured from the diseases (77.1%), the need for expensive drugs (63.9%), increased intensity and duration of the diseases (45.9%) and decreased antibiotic activity (45.2%) were reported as the consequences of AMR. The findings were higher than the findings in Kemssie, which were 28.6%, 20.0%, 22.6%, and 23.4%, respectively [ 24 ]. The difference might be due to the study setting, where Dessie is highly civilized and a metropolitan City compared to Kemssie.
In the current study, about 238 (58.5%, 95% CI: 53.1–63.1) respondents had good knowledge of AMR. The finding is nearly similar to the study done in Dire Dawa, Ethiopia (62.8%) [ 45 ], but higher than other studies conducted in Nigeria (8.3%) [ 31 ], and Pakistan (44.4%) [ 28 ]. The difference might be attributed to the difference in the questionnaire used, the criteria to say good knowledge and poor knowledge, the study setting and the health administration system employed in each country.
The result showed a lower level of knowledge of AMR among adults, which requires the attention of the stakeholders. Poor educational intervention about AMR among the public, poor focus of health professionals to advise about the risk of AMR, socio-demographic factors and poor attention of health administrators to AMR might be the reasons for lower levels of AMR knowledge [ 37 ]. Although overcoming knowledge deficits alone will be insufficient for global AMR behavior change, the authors believe education and awareness campaigns can increase the public’s knowledge of AMR [ 46 , 47 ].
Male gender, educational level, getting advice from health professionals about how to take antibiotics, using health professionals as a source of information on antibiotics, and taking antibiotics without a prescription were significantly associated with good knowledge of AMR.
Males were about 2 times more likely to have good knowledge than females. However, there was no association between the gender of respondents and knowledge of AMR in Nigeria [ 31 ]. This difference might be contextual (varied based on setting) and intersect with other socio-demographic factors, particularly education (in Ethiopia, females had lower educational level) and socioeconomic status (females had lower income level) [ 48 ]. As the educational level of the majority of females in the country is lower than male, it is likely that AMR knowledge is lower among females. The WHO recommended a need to have a gender and equity focus in all efforts because it is important to national efforts to tackle AMR [ 49 ].
In the current study, higher educational levels had a significant association with knowledge of AMR. Study participants with college and above educational levels had 3.5 times better knowledge of AMR and those with grade 11–12 educational levels had 3.7 times better knowledge of AMR than illiterate adults. This is in line with other studies [ 18 , 21 , 26 , 31 , 50 – 55 ]. The impact of educational training on improving the awareness, knowledge, and perception of AMR is very important [ 49 , 54 ].
The current study showed that participants who reported getting advice from health professionals about how to take antibiotics and using health professionals as sources of information on antibiotics had about 1.8 times and 2.5 times better knowledge than their counterparts, respectively. This is not surprising as health professionals are expected to hold better knowledge about antibiotics and sharing information with individuals could result in better knowledge of the respondents about AMR. Cascading WHO recommendations are crucial to address available information on population diversity to target and refine behavior change among the general population and/or certain occupations [ 49 ]. Study participants who ever took antibiotics without a prescription had 1.8 odds of better knowledge of AMR. This might be due to the experience of taking the medication leading to the acquiring of information through different ways like using the internet, medical books and other reading materials that could lead to enhancing knowledge of AMR.
Limitations of the study
Even though it could report many important findings, the current study had several limitations. Because it was a cross-sectional study, no cause and effect relationships could be established, and the sample size was small because we did not account for design effects. Controlling the difference between the two data collectors was difficult which might result in variations of the data. Even though the tool was used in the previous study in Ethiopia and we established its face validity, the content validity and reliability were not performed to assure tool validity in the current study.
The current study indicated poor awareness and knowledge of AMR among the adults in Dessie City. Being male, higher educational level, getting advice from health professionals about how to take antibiotics, using health professionals as a source of information on antibiotics, and taking antibiotics without a prescription were significantly associated with good knowledge of AMR. Educational campaigns based on socio-demographic contexts should be provided by the stakeholders to improve the awareness and knowledge of AMR among the public. As health professionals are potential sources of information on AMR, health administrators should plan to use them widely as mediators of information to the public, thereby increasing the awareness and knowledge of AMR among the public.
Supporting information
S1 table. socio-demographic characteristics of the study participants among adults in dessie city, northeast ethiopia, 2021 (n = 407)..
https://doi.org/10.1371/journal.pone.0279342.s001
S2 Table. Antibiotic use and related information among the adults of Dessie City, Northeast Ethiopia, 2021 (n = 407).
https://doi.org/10.1371/journal.pone.0279342.s002
S3 Table. Awareness and knowledge of AMR among the adults of Dessie City, Northeast Ethiopia, 2021 (n = 407).
https://doi.org/10.1371/journal.pone.0279342.s003
S4 Table. Association factors of knowledge towards AMR among the adults of Dessie City, Northeast Ethiopia, 2021 (n = 407).
https://doi.org/10.1371/journal.pone.0279342.s004
S5 Table. Annex 1: Questionnaires.
https://doi.org/10.1371/journal.pone.0279342.s005
S1 Dataset.
https://doi.org/10.1371/journal.pone.0279342.s006
Acknowledgments
The authors would like to acknowledge Mr. Degu Dagnew (Facilitator of the data collectors), Mr. Henok Dagne, Mr. Yigizie Yeshew, Wollo University, Dessie City administration and all study participants.
- View Article
- Google Scholar
- 2. Organization, WHO., Antimicrobial resistance and primary health care. 2018, World Health Organization. Availaibe at https://apps.who.int/iris/bitstream/handle/10665/326454/WHO-HIS-SDS-2018.56-eng.pdf .
- PubMed/NCBI
- 18. Antibiotic Resistance Multi-country public awareness survey. World Health Organization: Geneva, Switzerland, 2015: p. 59. https://apps.who.int/iris/handle/10665/194460 or https://apps.who.int/iris/bitstream/handle/10665/194460/9789241509817_eng.pdf?sequence=1&isAllowed=y .
- 37. WHO. Antibiotic Resistance:Multi-country public awareness survey 2015. https://apps.who.int/iris/bitstream/handle/10665/194460/9789241509817_eng.pdf?sequence=1&isAllowed=y .
- 49. World Health Organization. Tackling antimicrobial resistance (AMR) together: working paper 1.0: multisectoral coordination. 2018, World Health Organization. https://apps.who.int/iris/rest/bitstreams/1318162/retrieve .
- 52. Carter RR, Sun J, Jump RL. A survey and analysis of the American public’s perceptions and knowledge about antibiotic resistance. in Open Forum Infectious Diseases. Oxford University Press, 2016. https://doi.org/10.1093/ofid/ofw112 pmid:27382598.
PERSPECTIVE article
Crisis of antimicrobial resistance in china: now and the future.

- 1 Center of Infectious Disease, West China Hospital, Sichuan University, Chengdu, China
- 2 College of Pharmacy, University of Florida, Gainesville, FL, United States
The crisis of antimicrobial resistance is worsening and has become a major public safety problem in China, seriously endangering human and animal health and ecological environment. Gram-negative bacterial resistance in China is severe: the related pathogens mainly include carbapenem-resistant Acinetobacter, Pseudomonas aeruginosa and Klebsiella pneumoniae. Surging antimicrobial consumption and irrational use of antimicrobials are the main causes of resistance. In China, a variety of strategies are implemented to control the antimicrobial resistance in hospitals, agriculture and environment. However, there is still a long way to go to strengthen the drug resistance surveillance, to reduce the emergence of drug-resistant bacteria, and to find new antimicrobials and therapies for drug-resistant bacteria. Controlling the antimicrobial resistance crisis takes great efforts from the whole society.
Antimicrobials have saved tens of millions of lives since penicillin was used clinically in 1940s. They have made an outstanding contribution to prolong the average life span. However, many bacteria have developed severe resistance to antimicrobials with the increased antimicrobial consumption worldwide. The development rate of bacterial resistance is much faster than that of new antimicrobials. If uncontrolled, humans will enter the “post-antibiotic era.” China is one of the top consumers of antimicrobials in the world with 1.3 billion population. Therefore, it is more challenging for China to face the antimicrobial resistance.
The Resistance Pattern of Antimicrobials in China
Infections caused by multidrug-resistant organisms (MDROs), especially carbapenem-resistant Gram-negative bacteria often cause high mortality due to limited treatment options. Bacterial resistance data from multiple hospitals in China have been collected by China Antimicrobial Resistance Surveillance System (CARSS) ( National Health and Family Planning Commission of the People’s Republic of China, 2017 ). In 2016, a total of 2727605 strains from 1273 hospitals were collected. Imipenem-resistant Acinetobacter baumannii increased from 45.8% in 2012 to 59.2% in 2016. The resistance rate of Escherichia coli to imipenem and third-generation cephalosporins (3GC) decreased slightly from 2.2 and 69.7% in 2012 to 1.2 and 56.3% in 2016, respectively. From 2012 to 2016, the resistance rate of Klebsiella pneumoniae fluctuated with the rising trend, reaching 34.5 and 8.7% in 2016 to 3GC and carbapenems, respectively. In 2016, the prevalence of carbapenem-resistant Pseudomonas aeruginosa (CRPA), methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium was 22.3, 34.4, and 2.0% respectively. There was no S. aureus with vancomycin resistance. The China Antimicrobial Surveillance Network ( CHINET, 2018 ) began monitoring bacterial resistance nationwide since 2005. Their data helped us to understand the status and changes of bacterial resistance in China. Between 2005 and 2017, the number of bacterial strains isolated annually ranged between 22774 and 190610. Carbapenem-resistant Acinetobacter , of which over 90% were A. baumannii (CRAB), increased from 31 to 71.4%, with 60% being multidrug-resistant. Enterobacteriaceae were still highly sensitive to carbapenems, as the carbapenem resistance rate of most bacteria in this family was less than 10%. There was a slightly downward trend in the prevalence of P. aeruginosa and in its resistance rate to carbapenems (ranging from 20 to 30%, CRPA). Remarkably, there was a tenfold increase of the carbapenem-resistant K. pneumoniae (CRKP), from 2.4 to 24% in the past 13 years, most of which were isolated from sputum specimen. The geographical distribution of CRAB, CRKP and CRPA was mainly concentrated in central and eastern China and Yunnan Province ( CHINET, 2018 ). On the contrary, MRSA isolates decreased from 69% in 2005 to 35.3% in 2017. The prevalence of vancomycin-resistant Enterococci (VRE) in China is still low ( Hu et al., 2016 ; CHINET, 2018 ). In 2014, the first World Health Organization (WHO) global report on antimicrobial resistance surveillance showed that CRKP has appeared in almost all parts of the world ( World Health Organization [WHO], 2014 ). A report on antimicrobial resistance surveillance in Europe showed that the resistance rate of K. pneumoniae against carbapenems was 6.1% in 2016, without a significant change from 2013 to 2016. The prevalence of CRKP in Greece was the highest, up to 61.9% ( ECDC, 2016 ). In summary, just like the global trend, Gram-negative bacterial resistance in China is severe. In particular, the rapid growth of CRKP should draw widespread attention.
Drug resistance surveillance network for zoonotic bacteria in China was established in 2008 ( Zhang et al., 2017 ). The key monitored strains are E. coli, Salmonella, S. aureus, Enterococcus, Campylobacter, Streptococcus, Haemophilus parasuis , and Pasteurella , etc. The serotype identification and drug resistance testing have been completed in more than 30, 000 strains of bacteria. In recent years, bacteria such as E. coli and Salmonella carried by animals have been found to be resistant to colistin. The situation is worse in poultry than in swine. E. coli and Salmonella are also highly resistant to tetracycline. The resistance rate of E. coli to florfenicol has been as high as 100%, and to enrofloxacin about 50–70% ( Cai, 2017 ).
What Drives the Antimicrobial Resistance in China?
Some possible reasons for the increasing antimicrobial resistance in China are illustrated in Figure 1 .
Figure 1. Possible reasons for the increasing antimicrobial resistance in China. Possible reasons include increased antimicrobial consumption, irrational use of antimicrobials in clinical practice and agriculture, dissemination of ARGs with the increase of anthropogenic activities and more susceptible people for resistant organisms.
First, antimicrobial consumption promotes antimicrobial resistance. A study by Klein et al. (2018) showed that antimicrobial consumption increased by 79% [2.3–4.2 billion DDDs (defined daily doses)] in China between 2000 and 2015, higher than the increase of global antimicrobial consumption, which was 65% over the 15 years. The antimicrobial consumption rate in DDDs per 1,000 inhabitants per day in China (65%) also grew faster than that of the globe (39%). In China, the higher population density, the decreased air quality due to the emission of gasoline and other fuels in the urbanization, and the high prevalence of chronic obstructive pulmonary disease (COPD) (8.6%, 99.9 million people) ( Wang et al., 2018 ) make people become more susceptible to bacterial infections. In addition, more and more patients seek medical advice and have higher healthcare expectation with the continuous improvement of the social security system, living standards and health literacy. Therefore, the demand for antimicrobials has increased.
Secondly, irrational use of antimicrobial agents in clinical practice (especially among children) and agriculture (including livestock, aquatic products, and crops, etc.) ( Zeng et al., 2017 ). Some general practitioners or rural doctors are unfamiliar with the principles and methods for the rational application of antimicrobials, they may wrongly prescribe antimicrobials in the ways such as incorrect dosing, topical application of systemic antimicrobials, and improper antimicrobial prophylaxis, etc. Most often, patients with viral infections (flu or common cold) may be prescribed antimicrobials. A study from Poland also showed increased antimicrobial consumption in viral infection season ( Ciszewski et al., 2017 ). Financial incentives, such as mark-ups on drug price, is considered to be the main driver of over-prescribing in China ( Qiao et al., 2018 ). Many people have low literacy about antimicrobials, and they pursue antimicrobials through a pharmacy without prescription (online or on site) ( Wang et al., 2016 ). Antimicrobials are widely used in livestock as prophylactic and therapeutic agent for infections and as growth promoters. Antimicrobial use in livestock is even slightly higher than in humans ( Zhang et al., 2015 ). Antimicrobials have contaminated the food and drinking water supply in China because a large number of antimicrobials are used improperly in livestock in rural China ( Hao et al., 2015 ). In 2015, a survey on the antimicrobial body burden of Chinese schoolchildren found that 58.3% of 1064 urine samples were tested positive for antimicrobials, and that the contaminated environment and food may be the main sources of exposure ( Wang et al., 2015 ). This may have induced bacterial resistance and unbalanced flora distribution, damaged the immune function and nervous system, and produced other adverse drug reactions.
Thirdly, antibiotic resistance genes (ARGs) are a natural component of all environments. However, anthropogenic activities have led to the dissemination of ARGs as an emerging environmental contaminant ( Zhu et al., 2017 ; Qiao et al., 2018 ). Now, ARGs are widely distributed in China in environments including clinical areas, surface water, animal wastes, sewage treatment plant effluents and soils ( Qiao et al., 2018 ). Antimicrobials and ARGs can spread among the environment, humans and animals, which is closely associated with the increasing prevalence of antimicrobial resistance and is threatening human health. A recent study even found that smog metagenomes in Beijing contained multiple carbapenem-resistant genes. The relative abundance was similar to that in the human gut and sewage ( Pal et al., 2016 ).
Fourthly, bacterial defense dysfunction occurs in some populations as a result of the aging Chinese society, the long-term use of steroids and immunosuppressants, the extensive development of bone marrow and organ transplantation, the increased number of invasive treatments, and the prevalence of acquired immunodeficiency syndrome (AIDS). In these populations, their demand for antimicrobials is greater and they are more likely to become critically ill. They are more susceptible to resistant organisms because of risk factors including ICU admission, being elderly, indwelling devices (such as central venous catheters, catheters, endotracheal tubes) and invasive procedures ( Kaye and Pogue, 2015 ). It is very difficult to treat infections caused by multi-drug resistant strains in these patients. Maybe it is also one of the reasons for the severe bacterial resistance in China.
How to Deal With the Antimicrobial Resistance in China?
The theme for World Health Day 2011 was “combating drug resistance: no action today, no cure tomorrow,” which reinforced all countries around the world to take more active actions against bacterial resistance. The 2015 World Health Assembly adopted the global action on antimicrobial resistance. The 2016 UN High Level Meeting on antimicrobial resistance and the G20 Summit have made strong commitments to control antimicrobial resistance. What have we done? What else do we need to do?
The Chinese government attaches great importance to the issue of antimicrobial resistance and takes multiple measures to strengthen the antimicrobial stewardship. A series of documents were promulgated such as Administrative Measures for the Clinical Use of Antibacterial Drugs (2012) ( Ministry of Health of the PRC, 2012 ), Antimicrobial Management will be Enhanced in Multiareas (2015) ( National Health and Family Planning Commission of the PRC, 2015a ), Five Year Action Plan for the Comprehensive Management of Veterinary Drugs in China (2015–2019) ( Ministry of Agriculture and Rural Affairs of the PRC (2015–2019), 2015 ), National Action Plan to Contain Antimicrobial Resistance (2016-2020) ( National Health and Family Planning Commission of the PRC (2016–2020), 2016 ), and Work Program for the Reduction of the Use of Antimicrobials in Animals (2018–2021) ( Ministry of Agriculture and Rural Affairs of the People’s Republic of China (2018–2021), 2018 ), etc. These documents fully demonstrate that the state will strengthen supervision over the manufacture, circulation and use of antimicrobials and support the development of new antimicrobials. Therefore, regular training on rational use of antimicrobials, enhancement of antimicrobial stewardship, and strict implementation of infection control measures, especially hand hygiene have been carried out in the medical institutions at all levels to reduce the unnecessary consumption of antimicrobials and to delay the emergence and spread of resistant bacteria.
The management of clinical use of antimicrobials in China has gone through several stages: promulgation and implementation of guidelines for clinical use of antimicrobials (2004), specialized rectification (2011–2013), continuous improvement (2014–2017) with updated guidelines for clinical use of antimicrobials (2015), and refined and standardized management training since 2018 ( Ministry of Health of the PRC, 2004 ; National Health and Family Planning Commission of the PRC, 2011 , 2015b ; National Health Commission of the People’s Republic of China, 2018 ). These initiatives have achieved remarkable results through continuous efforts in recent years. According to the data from the Center for Antibacterial Surveillance ( National Health and Family Planning Commission of the People’s Republic of China, 2017 ), the use of antimicrobials in outpatient settings decreased from 17.2% in 2011 to 10.3% in 2016, in inpatient settings from 59.4% in 2011 to 37.5% in 2016. Antimicrobial drug use intensity decreased from 85.10 DDD in 2005 to 50.03 DDD in 2016. However, the sales of antimicrobials to children increased from ¥5 billion (US$ 781 million) in 2005 to ¥12 billion ($1.87 billion) in 2015 according to an analysis of Chinese children’s drug market in 2017. The National Health Commission issued the document on May 10, 2018 to strengthen the clinical application and management of antimicrobials for key populations such as children ( National Health Commission of the People’s Republic of China, 2018 ).
All the above shows that China’s firm determination to fight against bacterial resistance, but there are still many areas to be improved. Figure 2 is a schematic diagram. First, bacterial resistance surveillance in China is limited by its scope and uneven levels, especially antimicrobial resistance surveillance of animal-derived bacteria. Surveillance network of antimicrobial utilization and resistance patterns from pharmacies, clinics, hospitals, environment, agriculture and animal husbandry should be established at both local and national levels. Efforts should be made to strengthen the “big data” of bacterial resistance in all these sources, and to further address the intrinsic connections hidden behind the data. Nowadays, the development of whole-gene sequencing (WGS) technology can help researchers predict antimicrobial resistance more efficiently, thus assisting in clinical diagnosis and treatment decisions. WGS-powered online database, which developed by the Chinese scientists called BacWGSTdb, aims to pioneer the movement of WGS from proof-of-concept studies to routine use in clinical microbiology laboratory, offers a rapid and convenient platform to analyze epidemiological outbreak and the phylogeny of the bacterial genome, so as to provide information guarantee and decision-making support for prevention and control of infectious disease outbreaks and major bio-safety accidents ( Ruan and Feng, 2016 ; Quainoo et al., 2017 ; Rossen et al., 2018 ). Second, there is still irrational use of antimicrobials in healthcare (especially among children) and agriculture (especially in animal husbandry). The standards and regulations for environmental discharge of antimicrobials still need to be improved. Chinese government has launched some measures to halt financial incentives such as the separation of prescription sales from physicians’ income and the “zero mark-up” policy on drug sale. There is a huge amount of antimicrobial consumption in animal husbandry. Animal breeders, especially farmers, lack understanding of antimicrobial resistance and its hazards. Animal-use antimicrobials should be purchased for treatment with prescriptions from a veterinarian. Reducing the use of unnecessary antimicrobials in livestock farms has not significantly harmed animal health or farmers’ incomes ( Dierikx et al., 2016 ). Strengthen scientific breeding and management of livestock and poultry should help to reduce antimicrobial consumption in animals. Public awareness of the prevention and control of the bacterial resistance needs to be gradually raised. Third, it is necessary to continue to strictly control the sources of antimicrobial pollution from various aspects such as non-therapeutic use of antimicrobials and discharge of antimicrobial-containing sewage. The control of waste residue in antimicrobial industry should be taken into concern. Advanced oxidation processes (AOPs) might be used to improve the removal of ARGs in municipal sewage effluent ( Zhang et al., 2016 ). ARG metagenomic data make it possible to track ARG contamination sources ( Li et al., 2018 ), which is important for the control of ARG contamination. Fourth, we should actively explore the mode of antimicrobial stewardship suitable for the institutional development. Improve the organizational structure of antimicrobial clinical application, clarify the responsibilities and strengthen the fine management. Provide regular training and education for the rational application of antimicrobials. Establish a long-term mechanism of multidisciplinary case discussion and a multidisciplinary consulting team that consists of department of infectious diseases, clinical microbiological laboratory, department of pharmacy and department of nosocomial infection control. Under the support of information technology, strengthen the stratified management of antimicrobials, dynamically monitor the use of antimicrobial agents, evaluate the suitability and rectification of antimicrobial use. Transfer the management of antimicrobials from administrative intervention to multidisciplinary collaboration focusing on competency and patient-centered care. In addition, strengthen international cooperation is another important way to better implement antimicrobial stewardship and to stem the tide of antimicrobial resistance ( Hoffman et al., 2015 ). Fifth, at present, many policies and training of antimicrobial agents in China are concentrated in hospitals above the level-II, but about 60% of patients in the country are treated in community hospitals/rural hospitals (level-I). To tackle this mismatch, multi-disciplinary intervention methods such as professional training in the diagnosis and treatment of infectious diseases, use of social media tool such as WeChat and on-site training platforms organized by tertiary hospitals are offered to healthcare professionals and patients in community hospitals/rural hospitals to create a model for antimicrobial intervention in primary health care institutions in China. Sixth, active screening and enhanced interventions for MDRO’s colonization in high-risk patients should be an important way to reduce drug-resistant bacterial infections. Strict isolation enhanced environmental disinfection and hand hygiene should be implemented in MDRO-colonized patients. Currently, different de-planting measures have been adopted according to different colonizing bacteria and different planting sites. Oral cleaning is performed with chlorhexidine, nasal cleaning is performed with iodide or mupirocin: feces with intestinal drug-resistant bacteria are processed separately. Minimizing hospital length of stay and reducing the conversion rate from colonization to infections may be one of the most important infection control measures. Seventh, the scientific challenges are the main problems in the development of new antimicrobial agents. Low profit for pharmaceutical companies on research and development of new drugs is another obstacle. The use of antimicrobial peptides as adjuvants to antimicrobials could probably slow down the development of drug resistance ( Lázár et al., 2018 ). New research suggested that immunomimetic designer cells might be used to cure resistant bacterial infections in the future ( Liu et al., 2018 ). Promoting the development and use of vaccines may reduce the demand for antimicrobials, especially in animal feed. The rapid development of DNA sequencing and artificial intelligence (AI) makes it possible to screen phages rapidly and efficiently. Phage therapy is promising to be a powerful weapon against super drug-resistant bacteria. Eighth, there are trillions of bacteria and other microorganisms in the human body. They coexist in symbiosis with human beings. Many factors, such as irrational use of antimicrobials, unhealthy diet or long-term stress and anxiety could destroy the balance of microorganisms and lead to various diseases. This is consistent with the Traditional Chinese Medicine rationale “When there is sufficient healthy qi inside, pathogenic factors have no way to invade”. If people live in harmony with bacteria, fungi or viruses, people are less likely to get sick. In recent years, fecal microbiota transplantation (FMT) and probiotics are increasingly used in the treatment of various diseases such as Clostridium difficile infection, inflammatory bowel disease, constipation, diabetes and obesity, etc. ( Rossen et al., 2015 ). These methods are all based on the restoration of dysregulated intestinal flora to cure the diseases. Maybe it will provide new insights for reducing the emergence of resistant bacteria and finding new treatments in the future. Ninth, immunity dysfunction, especially immune suppression during late stage of infection plays an important role in the development and prognosis of severe infections ( Delano and Ward, 2016 ). If the immune status of patients with severe infections can be accurately monitored and effectively intervened, the prognosis of these patients could be significantly improved, then the dilemma of “no drug available” could be changed. There will be new discoveries and breakthroughs in the study of infection and immune balance.
Figure 2. Measures to be taken to combat antimicrobial resistance in China. Combating antimicrobial resistance, we need to carry out the work from community service and medical institutions. Community service includes strengthening bacterial resistance surveillance, reducing unnecessary antimicrobial use, controlling environmental pollution and intensify sewage treatment. Other measures that can be implemented in medical institutions include exploring the mode of antimicrobial stewardship, multidisciplinary antimicrobial consults in primary health care; screening and intervening multidrug-resistant organisms’ colonization; developing new antimicrobial drugs, peptides and phages; maintaining the balance of microorganisms in the human body and correcting immune imbalance at the right time.
In short, antimicrobial resistance in China must be curbed. It calls for the power of the whole society to control the antimicrobial resistance now and in the future. Antimicrobials are precious resources for us humans, and it is everyone’s responsibility to protect them.
Author Contributions
XL and JQ conceived and designed the study. JQ and YH wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
The authors would like to extend their sincere appreciation to Sichuan Province Science and Technology Support Program of China for funding this research work (No. 2017SZ0140).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Cai, X. P. (2017). Strengthen the management of antimicrobials, curb resistance of animal derived bacteria [Chinese]. Vet. Orientat. 23, 6–8.
Google Scholar
CHINET (2018). Institute of Antibiotics, Huashan Hospital, Fudan University. Available at: http://www.chinets.com/ (accessed September 29, 2018).
Ciszewski, M., Czekaj, T., and Szewczyk, E. M. (2017). Outpatient antibiotic consumption fluctuations in a view of unreasonable antibacterial therapy. Pol. J. Microbiol. 66, 119–123. doi: 10.5604/17331331.1235000
PubMed Abstract | CrossRef Full Text | Google Scholar
Delano, M. J., and Ward, P. A. (2016). Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J. Clin. Invest. 126, 23–31. doi: 10.1172/JCI82224
Dierikx, C. M., Hengeveld, P. D., Veldman, K. T., de Haan, A., van der Voorde, S., Dop, P. Y., et al. (2016). Ten years later: still a high prevalence of MRSA in slaughter pigs despite a significant reductionin antimicrobial usage in pigs the Netherlands. J. Antimicrob. Chemother. 71, 2414–2418. doi: 10.1093/jac/dkw190
ECDC (2016). Surveillance of Antimicrobial Resistance in Europe. https://ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2016 (accessed September 29, 2018).
Hao, R., Zhao, R., Qiu, S., Wang, L., and Song, H. (2015). Antibiotics crisis in China. Science 348, 1100–1101. doi: 10.1126/science.348.6239.1100-d
Hoffman, S. J., Caleo, G. M., Daulaire, N., Elbe, S., Matsoso, P., Mossialos, E., et al. (2015). Strategies for achieving global collective action on antimicrobial resistance. Bull. World Health Organ. 93, 867–876. doi: 10.2471/BLT.15.153171
Hu, F. P., Guo, Y., Zhu, D. M., Wang, F., Jiang, X. F., Xu, Y. C., et al. (2016). Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin. Microbiol. Infect. 22(Suppl. 1), S9–S14. doi: 10.1016/j.cmi.2016.01.001
Kaye, K. S., and Pogue, J. M. (2015). Infections caused by resistant Gram-negative bacteria: epidemiology and management. Pharmacotherapy 35, 949–962. doi: 10.1002/phar.1636
Klein, E. Y., Van Boeckel, T. P., Martinez, E. M., Pant, S., Gandra, S., Levin, S. A., et al. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U.S.A. 115, E3463–E3470. doi: 10.1073/pnas.1717295115
Lázár, V., Martins, A., Spohn, R., Daruka, L., Grézal, G., Fekete, G., et al. (2018). Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731. doi: 10.1038/s41564-018-0164-160
Li, L. G., Yin, X., and Zhang, T. (2018). Tracking antibiotic resistance gene pollution from different sources using machine-learning classification. Microbiome 6:93. doi: 10.1186/s40168-018-0480-x
Liu, Y., Bai, P., Woischnig, A. K., Charpin-El, Hamri G, Ye, H., Folcher, M., et al. (2018). Immunomimetic Designer Cells Protect Mice from MRSA Infection. Cell 174, 259–270. doi: 10.1016/j.cell.2018.05.039
Ministry of Agriculture and Rural Affairs of the People’s Republic of China (2018–2021) (2018). Work Program for the Reduction of the Use of Antimicrobials in Animals. Beijing: Ministry of Agriculture and Rural Affairs of the People’s Republic of China.
Ministry of Agriculture and Rural Affairs of the PRC (2015–2019) (2015). Five Year Action Plan for the Comprehensive Management of Veterinary Drugs in China. Beijing: Ministry of Agriculture and Rural Affairs of the PRC.
Ministry of Health of the PRC (2004). Guiding Principles of Clinical Use of Antibiotics. Beijing: Ministry of Health of the PRC.
Ministry of Health of the PRC (2012). Administrative Measures for the Clinical Use of Antibacterial Drug. Beijing: Ministry of Health of the PRC.
National Health and Family Planning Commission of the People’s Republic of China (2017). Report on Current Status of Antimicrobial Agent Management and Antimicrobial Resistance in China. Beijing: Beijing Union Medical University Press.
National Health and Family Planning Commission of the PRC (2016–2020) (2016). National Action Plan to Contain the Resistant of Antimicrobial Resistance. Beijing: National Health and Family Planning Commission of the PRC.
National Health Commission of the People’s Republic of China (2018). Continuously Manage the Clinical Application of Antimicrobial agents [Internet]. Beijing: National Health Commission of the People’s Republic of China.
National Health and Family Planning Commission of the PRC (2011). Special Rectification of Antibacterial drug Clinical Application. Beijing: National Health and Family Planning Commission of the PRC.
National Health and Family Planning Commission of the PRC (2015a). Antimicrobial Management will be Enhanced in Multiareas. Beijing: National Health and Family Planning Commission of the PRC.
National Health and Family Planning Commission of the PRC (2015b). Guiding Principles of Clinical Use of Antibiotics. Beijing: National Health and Family Planning Commission of the PRC.
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. (2016). The structure and diversity of human, animal and environmental resistomes. Microbiome 4:54.
PubMed Abstract | Google Scholar
Qiao, M., Ying, G. G., Singer, A. C., and Zhu, Y. G. (2018). Review of antibiotic resistance in China and its environment. Environ. Int. 110, 160–172. doi: 10.1016/j.envint.2017.10.016
Quainoo, S., Coolen, J. P., van Hijum, S. A., Huynen, M. A., Melchers, W. J., van Schaik, W., et al. (2017). Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 30, 1015–1063. doi: 10.1128/CMR.00016-17
Rossen, J. W., Friedrich, A. W., Moran-Gilad, J., ESCMID Study Group for Genomic, and Molecular Diagnostics (ESGMD), (2018). Practical issues in implementing. (whole)-genome-sequencing in routine diagnostic microbiology. Clin. Microbiol. Infect. 24, 355–360. doi: 10.1016/j.cmi.2017.11.001
Rossen, N. G., MacDonald, J. K., de Vries, E. M., D’Haens, G. R., de Vos, W. M., Zoetendal, E. G., et al. (2015). Fecal microbiota transplantation as novel therapy in gastroenterology: a systematic review. World J. Gastroenterol. 21, 5359–5371. doi: 10.3748/wjg.v21.i17.5359
Ruan, Z., and Feng, Y. (2016). BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 44, D682–D687. doi: 10.1093/nar/gkv1004
Wang, C., Xu, J., Yang, L., Xu, Y., Zhang, X., Bai, C., et al. (2018). Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): a national cross-sectional study. Lancet 391, 1706–1717. doi: 10.1016/S0140-6736(18)30841-30849
Wang, H. X., Wang, B., Zhao, Q., Zhao, Y., Fu, C., Feng, X., et al. (2015). Antibiotic body burden of Chinese school children: a multisite biomonitoring-based study. Environ. Sci. Technol. 49, 5070–5079. doi: 10.1021/es5059428
Wang, L., Zhang, X., Liang, X., and Bloom, G. (2016). Addressing antimicrobial resistance in China: policy implementation in a complex context. Global. Health 12:30. doi: 10.1186/s12992-016-0167-167
World Health Organization [WHO] (2014). Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization.
Zeng, L., Hu, D., Choonara, I., Mu, D., Zhang, L., Li, X., et al. (2017). A prospective study of the use of antibiotics in the emergency department of a Chinese university hospital. Int. J. Pharm. Pract. 25, 89–92. doi: 10.1111/ijpp.12335
Zhang, C. P., Song, L., Wu, C. B., and Xu, S. X. (2017). Drug resistance surveillance network for zoonotic bacteria in china [Chinese]. China Anim. Health Inspect. 34, 34–38.
Zhang, Q. Q., Ying, G. G., Pan, C. G., Liu, Y. S., and Zhao, J. L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 49, 6772–6782. doi: 10.1021/acs.est.5b00729
Zhang, Y. Y., Zhuang, Y., Geng, J. J., Ren, H. Q., Xu, K., and Ding, L. L. (2016). Reduction of antibiotic resistance genes in municipal wastewater effluent by advanced oxidation processes. Sci. Total Environ. 15, 184–191. doi: 10.1016/j.scitotenv.2016.01.078
Zhu, Y. G., Zhao, Y., Li, B., Huang, C. L., Zhang, S. Y., Yu, S., et al. (2017). Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2:16270. doi: 10.1038/nmicrobiol.2016.270
Keywords : crisis, antimicrobial resistance, China, resistance pattern, combating drug resistance
Citation: Qu J, Huang Y and Lv X (2019) Crisis of Antimicrobial Resistance in China: Now and the Future. Front. Microbiol. 10:2240. doi: 10.3389/fmicb.2019.02240
Received: 02 April 2019; Accepted: 12 September 2019; Published: 27 September 2019.
Reviewed by:
Copyright © 2019 Qu, Huang and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoju Lv, [email protected]
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- v.13; 2021 Dec
Antimicrobial resistance in wildlife and in the built environment in a wildlife rehabilitation center
Carla baros jorquera.
a Escuela de Medicina Veterinaria, Facultad Ciencias de la Vida, Universidad Andrés Bello. Av. República 440, Santiago, Chile
Andrea I. Moreno-Switt
b Escuela de Medicina Veterinaria, Facultad de Agronomía e Ingeniería Forestal, Facultad de Ciencias Biológicas, Facultad de Medicina, Pontificia Universidad Católica de Chile. Av. Vicuña Mackenna 4860 Macul, Santiago, Chile
c Millennium Initiative for Collaborative Research On Bacterial Resistance (MICROB-R). Av. Las Condes 12.438, Lo Barnechea, Santiago, Chile
Nicole Sallaberry-Pincheira
d Unidad de Rehabilitación de Fauna Silvestre, Escuela de Medicina Veterinaria, Facultad de Ciencias de la Vida, Universidad Andrés Bello, Av. República 440, Santiago, Chile
Jose M. Munita
e Genomics and Resistant Microbes Group, Facultad de Medicina Clínica Alemana, Universidad del Desarrollo, Av. Las Condes 12.438, Lo Barnechea, Santiago, Chile
Camila Flores Navarro
Rodolfo tardone, gerardo gonzález-rocha.
f Laboratorio de Investigación en Agentes Antibacterianos, Facultad de Ciencias Biológicas, Universidad de Concepción, Concepción, Chile
Randall S. Singer
g Department of Veterinary and Biomedical Sciences, College of Veterinary Medicine, University of Minnesota, 1971 Commonwealth Avenue, St. Paul, MN 55108. United States.
Irene Bueno
Associated data.
Injured and orphaned wildlife are often brought to Wildlife Rehabilitation Centers (WRC) to be cared for by professionals to ultimately be released back to their natural habitats. In these centers, animals may spend months and frequently receive prolonged antibiotic therapy. Therefore, WRC may play a role in the emergence and dissemination of antimicrobial resistance (AMR). The goal of this study was to investigate the presence and antibiotic resistance profiles of Gram-negative bacteria with reduced susceptibility to cephalosporins in both the wildlife admitted to a WRC and in the WRC built environment in Chile. A cross-sectional study was conducted sampling animals undergoing rehabilitation ( n = 64) and the WRC environment ( n = 160). Isolated bacterial species were identified with MALDI-TOF, and antimicrobial susceptibility determined using the disk diffusion method. Enterobacteriaceae and Pseudomonadaceae were the dominant bacterial families among the environmental ( n = 78) and animal ( n = 31) isolates. For Enterobacteriaceae , isolates of the most abundant species ( E. coli ) were classified into 20 antibiotic resistance profiles, with eight of those isolates being resistant to more than nine antibiotics, including imipenem. Isolates of the Pseudomonadaceae family identified 11 isolates with resistance to antibiotics such as carbapenems and quinolones. Even though a cluster analysis based on antibiotic resistance patterns did not show a clear overlap between environmental and animal isolates, it is important to highlight the identification of isolates resistant to carbapenems, which is very relevant from a public health perspective. Further, numerous antibiotic resistance profiles were observed in different bacterial species, indicating not only environmental contamination with a wide diversity of bacteria, but also a wide diversity of resistant bacteria in animals at the WRC. The approach taken by sampling animals and their hospital environment can be useful in understanding AMR dynamics in wildlife rehabilitation settings, as well as the potential dissemination of AMR into the natural environment.
- • The wildlife center was contaminated with wide diversity of resistant bacteria.
- • There was wide diversity of resistant bacteria in wildlife at the center.
- • Resistant isolates to carbapenems were present, which has public health relevance.
- • No clear overlap between wildlife and the center antibiotic resistance patterns.
- • Wildlife rehabilitation should be considered in antimicrobial resistance dynamics.
1. Introduction
The interconnectedness between humans, animals, and the natural environment (otherwise known as One Health) is key in understanding and mitigating antimicrobial resistance (AMR) given that resistant bacteria and resistance genes have the ability to move between these three compartments [ [1] , [2] , [3] ]. Of these three compartments, the role of the natural environment ( e.g. , soil, water, air, and wildlife) in the ecology and dissemination of AMR has received increased attention and has been reviewed in several recent publications [ 1 , [4] , [5] , [6] ]. Waste from anthropogenic sources, such as hospitals, wastewater treatment plants, pharmaceutical industries, and agricultural activities are ultimately released into natural environments. This waste may contain antibiotics, their metabolites, antibiotic resistant bacteria, and resistance genes. Thus, the natural environment may act as a reservoir and as a pathway of AMR spread to humans, animals, and the natural ecosystem [ 4 , 7 ].
AMR is a phenomenon that has existed for eons, well before the ‘antibiotics era’. This has been shown in studies where antibiotic resistant bacteria and/or antibiotic resistance genes usually found in clinical settings have been detected in areas far-removed from human contact [ 8 , 9 ]. Despite it being a natural phenomenon, anthropogenic pressures, such as human wastewater systems or animal husbandry facilities, may increase the occurrence, diversity, and quantity of antibiotic resistant bacteria and genes in the environment [ 10 , 11 ].
Wildlife species are part of the natural environmental compartment and can also naturally harbor antibiotic resistant bacteria. However, selective and anthropogenic pressures may also increase the potential for free-ranging wildlife to carry emerging resistant bacteria and genes, as well as facilitate their dissemination [ [12] , [13] , [14] ]. Injured and orphaned wildlife are often brought to wildlife rehabilitation centers (WRC) so that they can be cared for by professionals to ultimately be released back to their natural habitats. In these centers, animals may spend months and frequently receive prolonged antibiotic therapy [ 15 ]. There are studies that have reported the presence of antibiotic resistant bacteria and resistance genes, including those of public health concern, in wild animals admitted to WRC in different parts of the world. Giacopello et al. (2016) found multi-drug resistant Enterobacteriaceae (resistant to three or more antibiotics) in wild birds admitted to a rehabilitation center in Italy [ 16 ]. In another study, Darwich et al. (2019) detected bacterial isolates resistant to fluoroquinolones, tetracyclines and aminoglycosides (among others), and cephalosporin resistant genes from wildlife admitted to a rehabilitation center in Spain [ 17 ]. Within Chile, antibiotic resistant bacteria and genes, including those of public health relevance, have been found in wildlife admitted to WRC. Specifically, extended spectrum beta-lactamases (ESBL)-producing Escherichia coli and Salmonella enterica serovar Infantis were found in wild owls [ 13 ], and mec A and bla CTX-M genes were found in Andean foxes ( Lycalopex culpaeus ). These studies however sampled only the animals but not the hospital environment where they were housed. These studies illustrate the importance of not only evaluating the role of free-ranging wildlife but also the role that WRC have in the epidemiology of AMR emergence and spread, especially as the number of injured wild animals continues to increase due to a growing number of human-wildlife interactions [ 18 , 19 ].
The goal of this study was to investigate antibiotic resistance profiles of Gram-negative bacteria with reduced susceptibility to cephalosporins in both the wildlife species admitted to a WRC in Chile and in the WRC hospital built environment [ 20 , 21 ]. We hypothesized that Gram negative antibiotic resistant bacteria are widespread in the WRC built environment and that antibiotic resistance profiles recovered from animals hospitalized at the center would be similar to those observed in the WRC built environment.
2. Materials and methods
2.1. sampling design.
A cross-sectional study was conducted at the Wildlife Rehabilitation Center at the Universidad Andrés Bello (UFAS), located in the city of Santiago, Metropolitan Region of Chile. The center receives an average of 600 animals per year of different species of mammals, birds, reptiles, and amphibians. The main causes of admission to the WRC are wildlife attacked by domestic carnivores, vehicle collisions, illegal hunting, illegal wildlife trafficking and/or possession, and intoxication. Animals are mostly received from the Metropolitan Region of Chile, but a smaller number of them are admitted from other regions of the country as well.
The WRC is comprised of the following sectors (and subdivisions): reception, kitchen, quarantine, exam room, hospital (hospital 1 and hospital 2), indoor (indoor 1, 2, and 3), outdoor (outdoor 1, 2, and 3), and soft release (small animal enclosure, semi-aquatic bird enclosure, small bird enclosure, carnivore enclosure, flight room, owl enclosure, and parrot aviary). The specific number and type of samples taken per sector and subdivision can be found in Table 1 . In total, 160 samples at the WRC environment were collected with a gauze previously enriched in peptonized water in 100 mL sterile containers and passed through a 30 cm 2 sampled surface.
Total number of environmental samples ( n = 160) and number of ceph-resistant isolates that were taken from each sector, subdivision, and equipment (when applicable) at the wildlife rehabilitation center. The numbers represent the sample size and the percentage (%) from the total.
NA: Not applicable.
A random sample of animals from each sector of the rehabilitation center that were hospitalized on the day of the study were selected for sampling ( n = 64). This not only included animals from each sector, but also undergoing different stages of the rehabilitation process, as well as different taxa, to have a good representative cross-sectional sample of the animals at the WRC ( Table 2 ). Experienced veterinarians and trained volunteers collected rectal and/or cloacal swabs using a Cary Blair transport medium (Deltalab, Spain). In addition, data about the animals sampled (species, gender, age, animal admission date, origin, cause of admission, and previous antimicrobial therapy consisting of antibiotics used and length of treatment) were collected when available. All samples (environmental and animal) were kept at 4 °C until further analysis at the Universidad Andrés Bello research laboratory, where they were processed within 8 h of collection. The study was approved by the Universidad Andrés Bello bioethics committee (Act. 019/2014).
Summary table for the animal samples ( n = 64) with numbers and percentage (%) for each category, and number of ceph-resistant isolates.
2.2. Laboratory methods
For environmental samples, sterile containers with peptone water and gauzes were subjected to mixing by pulse vortexing for 15 s; this was followed by streaking 50 μL onto MacConkey agar (Becton Dickinson GmbH, Germany) supplemented with 1 mg/L of cefotaxime (Merck, Germany), as previously described [ 20 , 21 ]. For the animal samples, swabs were directly streaked into MacConkey agar, supplemented with cefotaxime as described above. All plates were incubated for 24–48 h at 37 °C, as previously described [ 22 ]. After incubation, distinct morphotypes were further isolated with at least three passages, and then isolated colonies were stored at −80 °C with 20% of glycerol.
Species identification was performed using a Vitek MS MALDI-TOF (bioMerieux, San Louis, MO, USA) following manufacturer's instructions as previously described [ 23 ]. Their antibiotic susceptibility profile was assessed using the disk diffusion method as per The Clinical & Laboratory Standards Institute (CLSI) recommendations [ 24 ]. Briefly, isolates were grown overnight in Tryptic Soy Broth (Becton Dickinson GmbH, Germany), then cultures were adjusted to a MacFarland 0.5 [ 25 ] and streaked in Muller Hilton agar (Becton Dickinson GmbH, Germany). All colonies representing different morphotypes that were grown on cephalosporin supplemented MacConkey Agar were further species identified and classified into families: Enterobacteriaceae/Yersiniaceae (order Enterobacteriales), Pseudomonadaceae, Comamonadaceae, Moraxellaceae, Xanthomonadaceae, and Alcaligenaceae. The combination of antibiotics tested varied according to bacterial families: Enterobacteriaceae/Yersiniaceae (order Enterobacteriales ), Pseudomonadaceae , Comamonadaceae , Moraxellaceae , Xanthomonadaceae , and Alcaligenaceae . CLSI breakpoints were used to characterize the antibiotic resistance patterns [ 24 ]. For Enterobacteriales, 19 antibiotics were tested: amikacin (AMK), gentamicin (GEN), ampicillin (AMP), amoxicillin/clavulanic acid (AMC), ampicillin/sulbactam (SAM), piperacillin/tazobactam (TZP), cefazolin (CFZ), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), chloramphenicol (CHL), ciprofloxacin (CIP), fosfomycin (FOS), tetracycline (TET), and trimethoprim/sulfamethoxazole (SXT). For Pseudomonadaceae , eight antibiotics were tested: AMK, GEN, CAZ, FEP, IPM, MEM, CIP, and TZP. For Moraxellaceae , 10 antibiotics were tested: AMK, GEN, SAM, TZP, CAZ, FEP, IPM, MEM, CIP, and SXT. For Xanthomonadaceae , three antibiotics were tested: CAZ, levofloxacin (LEV), and SXT. Finally, CAZ, MEM, and SXT were tested for Comamonadaceae and Alcaligenaceae . All disks were obtained from OXOID, United Kingdom. The control strain Escherichia coli ATCC25922 was used. The inhibition zone diameters were interpreted following the Susceptible, Intermediate, Resistant (SIR) status from the Clinical and Laboratory Standards Institute guidelines [ 24 ], which differed depending on the bacterial family and species (Tables A.1-A.5).
2.3. Data analyses
Antibiotic resistance patterns for both environmental and animal samples were described for each bacterial family. Further analyses focused on Enterobacteriaceae/Yersiniaceae (order Enterobacteriales ) and Pseudomonadaceae as most isolates belonged to these families. Fisher exact test was used to compare the frequency of isolates from animals with a history of previous antimicrobial exposure (yes/no) and their antibiotic resistance outcome (susceptible/intermediate/resistant) for Enterobacteriales order and for Pseudomonadaceae family separately across all the antibiotics tested. Statistical significance was defined with an alpha level of 5%.
A cluster analysis was performed to describe the resistance patterns obtained from Enterobacteriales and Pseudomonadaceae . The goal of the cluster analysis was to determine if isolates from the animals and the WRC environment were similar in their resistance profiles, as evidenced by isolates from both sources clustering together. To perform the cluster analysis, the zone of inhibition obtained for each isolate/antibiotic combination was used. Isolates susceptible to all antibiotics were removed prior to the analysis. Agglomerative Hierarchical Clustering (HC) was used, which is based on a dissimilarity matrix and has the advantage of not having the number of clusters chosen a priori [ 26 ]. The Gower distance was used to calculate the distance matrix, and Ward's method was used as the HC algorithm [ 27 ]. The functions ‘hclust’ and ‘daisy’ from the package ‘cluster’ in R were used to conduct the HC and the Gower distance respectively [ 28 ]. The optimal number of clusters was validated using the optimum average silhouette width with the ‘pamk’ function from the ‘fpc’ package in R [ 29 ]. All statistical analyses were performed in R software 3.6.3 [ 30 ].
3.1. Presence of different families of resistant bacteria in the environmental samples
A total of 160 samples were collected from the WRC environment and a total of 78 bacterial isolates were recovered ( Table 1 ). While isolates were obtained from all sampled sectors, most isolates were retrieved from the hospitals ( n = 28), quarantine ( n = 23), and soft release ( n = 15) ( Table 1 ). Further identification demonstrated that these isolates belonged to six bacterial families: 62.3% ( n = 48) Pseudomonadaceae , 21.8% ( n = 17) Enterobacteriaceae , 11.5% ( n = 9) Yersiniaceae , 1.3% (n = 1) Alcaligenaceae , 2.6% (n = 1) Moraxellaceae , and 1.3% (n = 1) Xanthomonadaceae . For Enterobacteriaceae , species identified were Citrobacter braakii, Escherichia coli, E. vulneris, and Enterobacter cloacae . For Yersiniaceae , Rahnella aquatilis . For Pseudomonadaceae , species identified were P. aeruginosa, P. fluorescens , P. oryzihabitans, P. putida , P. stutzeri , and P. viridiflava . For Alcaligenaceae , Achromobacter xylosoxidans was identified. For Moraxellaceae, Acinetobacter baumannii , and for Xanthomonadaceae, Stenotrophomonas maltophilia was identified.
3.2. Presence of different families of resistant bacteria in the animal samples
A total of 64 animal samples were collected. Of those, 86.0% ( n = 55) were avian species, 3.0% ( n = 2) mammals, and 11.0% ( n = 7) reptiles. There was a total of 25 different animal species, with the most common being Falco sparverius (n = 7), Milvago chimango (n = 7), and Tyto alba ( n = 6) for birds, Chelonoidis chilensis (n = 5) for reptiles, and Lycalopex culpaeus (n = 2) for mammals ( Table 2 ). Across all taxa, 54.7% ( n = 35) were adults, 28.1% ( n = 18) were juveniles, 15.6% ( n = 10) were nestlings/pups, and in 1.6% (n = 1) age was not determined. In 64.1% ( n = 41) of animals, gender was not determined, and for those where gender was determined, 18.6% ( n = 12) were female and 17.2% ( n = 11) were male. There was no information about the geographical origin of the animals for most animals sampled (60%, n = 38). For those with information about recovery location, the most frequent were counties 40-50 km from the city of Santiago. The average length of stay at the WRC among animals sampled was 6.7 months (range: 1 week - 3 years).
A total of 31 bacterial isolates were recovered from animal samples. These isolates were obtained from six different bird species: Turdus falcklandii ( n = 5), Tyto alba ( n = 4), Phrygilus fruticeti ( n = 3), Spatula platalea ( n = 2), Veniliornis lignarius ( n = 2), and Vanellus chilensis ( n = 1); one mammal species (three isolates from Lycalopex culpaeus ), and two reptile species ( Chelonoidis chilensis [ n = 9] and Philodryas chamissonis [n = 2]).
These 31 isolates were further classified into five bacterial families: 51.6% ( n = 16) Enterobacteriaceae , 29.0% (n = 9) Pseudomonadaceae , 9.7% (n = 3) Xanthomonadaceae , 6.5% (n = 2) Comamonadaceae , and 3.2% (n = 1) Moraxellaceae . For Enterobacteriaceae , Citrobacter braakii , Escherichia coli, E. vulneris, and Enterobacter cloacae were identified . For Pseudomonadaceae , Pseudomonas aeruginosa , P. deovorans , P. fluorescens , P. oryzihabitans , P. putida , P. stutzeri, and P. viridiflava. Stenotrophomonas maltophilia was the species identified for Xanthomonadaceae , Comamonas aquatica for Comamonadaceae , and Acinetobacter baumanni complex for Moraxellaceae .
3.3. Antimicrobial resistance in isolated bacteria from the environment and animals
For the order Enterobacteriales, resistance to antibiotics of different classes was found. Isolates obtained from environmental samples were resistant to penicillins (100%), cephalosporines (92.3%), aminoglycosides (42.3%), quinolones (42.3%), tetracyclines (38.4%), sulfonamides (30.8%), Fosfomycin (30.8%), chloramphenicol (23.1%), and carbapenems (11.5%). Isolates obtained from animal samples were resistant to penicillins (100%), cephalosporines (100%), tetracyclines (75.0%), quinolones (62.5%), sulfonamides (50%), chloramphenicol (31.3%), carbapenems (6.2%), and aminoglycosides (6.2%).
Numerous resistance profiles were found that further characterized the collected isolates. Isolates of the most abundant species ( E. coli ) were found in the environment and animal samples, and these isolates were classified into 20 antibiotic resistance profiles ( Table 3 ). The majority of E. coli isolates were resistant to more than 9/19 antibiotics tested, including highly resistant isolates, with one E. coli isolate from a sample obtained in the quarantine room that was resistant to 11/19 antibiotics (AMP-SAM-CFZ-FEP-CRO-FOX-ETP-IPM-MEM-GEN-FOS). One E. coli isolate from a sample obtained from a L. culpaeus also showed resistance to 11/19 antibiotics (AMP-SAM-AMC-CFZ-FEP-CRO-FOX-CAZ-TET-CIP-SXT).
Antimicrobial resistance profiles identified in isolates of the order Enterobacteriales and of the Pseudomonadaceae and Xanthomonadaceae families.
Abbreviations: amikacin (AMK), gentamicin (GEN), ampicillin (AMP), amoxicillin/clavulanic acid (AMC), ampicillin/sulbactam (SAM), piperacillin/tazobactam (TZP), cefazolin (CFZ), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), chloramphenicol (CHL), ciprofloxacin (CIP), fosfomycin (FOS), tetracycline (TET), levofloxacin (LEV), and trimethoprim/sulfamethoxazole (SXT).
Isolates of the Pseudomonadaceae family were tested using 8 antibiotics , which further identified 11 isolates with resistance to antibiotics such as carbapenems and quinolones, including four isolates collected from the environment and animals ( Table 3 ). For instance, one isolate of Pseudomonas aeruginosa obtained from an animal possessed resistance to TZP-GEN-CIP and another from the environment possessed resistance to IPM-MEM. Other species of Pseudomonas also possessed resistance to IPM-MEM ( Table 3 ).
In addition, one isolate from an environmental sample from the family Moraxellaceae, was identified as Acinetobacter baumannii and was susceptible to all 10 antibiotics. For Xanthomonadaceae , one environmental isolate and one animal isolate of Stenotrophomonas maltophilia were resistant to one of the three antibiotics tested and to the three antibiotics respectively ( Table 3 ). Lastly, for Alcaligenaceae there was one species identified, Achomobacter xylosoxidans, which was susceptible to all three antibiotics tested.
3.4. Antibiotic use, antibiotic resistance, and clustering
Out of the animals sampled, 31.2% ( n = 20) had received antibiotics (at least one dose of one antibiotic at some point in time) during their stay at the rehabilitation center, and 68.8% ( n = 44) had not. Clindamycin ( n = 10) followed by enrofloxacin ( n = 8) were the most commonly used antibiotics. The longest antibiotic treatment was 3.9 months for enrofloxacin in a Patagonian land turtle ( C. chilensis ), and the shortest antibiotic treatment was a seven-day course of enrofloxacin in an Andean fox ( L. culpaeus ) .
The 16 Enterobacteriaceae isolates that were recovered from animal samples belonged to 10 different animal species of which five had received antibiotic treatment and five had not. The nine Pseudomonadaceae isolates belonged to eight different animals of which one had received antibiotics and the remaining seven had not. There was no difference in the frequency of resistant isolates regardless of whether they had received antibiotics or not for Enterobacteriaceae ( p = 0.35) and for Pseudomonadaceae ( p = 0.56).
For the cluster analyses, 42 isolates for the order Enterobacteriales (26 from environmental samples and 16 from animal samples) and 11 for Pseudomonadaceae (8 for environmental samples and 3 for animal samples) were analyzed. The optimal number of clusters was two for Enterobacteriales (cluster I with 30 isolates and cluster II 12 isolates), and four for Pseudomonadaceae (cluster I with 4 isolates, cluster II with 4 isolates, cluster III with 2 isolates, and cluster IV with one isolate). In Enterobacteriales, cluster I isolates were resistant to 42.1% (8/19) of antibiotics, while isolates in cluster II were resistant to 15.8% (3/19) of antibiotics ( Table 4 ). Cluster I was dominated by small groupings of isolates obtained from C. chilensis , Tyto alba , and hospital 1 isolates, while cluster II only contained environmental isolates that belonged mostly to the kitchen, owl enclosure, and hospital 2 ( Fig. 1 ). For Pseudomonadaceae , Clusters II and III only contained environmental isolates and were dominated by hospital 2 and quarantine isolates, Cluster I had a mixed of animal and environmental isolates, and Cluster IV was made of an isolate of Vanelus chilensis ( Table 5 , Fig. 2 ).
Cluster results for Enterobacteriales isolates. The numbers represent the mean inhibition zone diameters in mm for each antibiotic that was tested. The number of isolates for each cluster is divided between environmental and animal isolates. Cluster I: 14 environmental and 16 animal isolates; Cluster II: 12 environmental isolates. The greyed-out fields represent those that are resistant according to the CLSI Susceptible Intermediate Resistant (SIR) status [ 24 ].
Abbreviations: AMK: amikacin; GEN: gentamicin; AMP: ampicillin; AMC: amoxicillin/clavulanic acid; SAM: ampicillin/sulbactam; TZP: piperacillin/tazobactam; CFZ: cefazolin; FOX: cefoxitin; CAZ: ceftazidime; CRO: ceftriaxone; FEP: cefepime; ETP: ertapenem; IPM: imipenem; MEM: meropenem; CHL: chloramphenicol; CIP: ciprofloxacin; FOS: fosfomycin; TET: tetracycline; SXT: sulphamethoxazole/trimethoprim.

Dendrogram for Enterobacteriales that resulted from the cluster analysis. The y-axis (height) represents how close together observations were when they were merged into clusters. gower_distR refers to Gower distance which was used to calculate the distance matrix, and Ward's refers to the method used as the hierarchical clustering algorithm. The rectangular boxes represent each one of the two clusters (I and II).
Cluster results for Pseudomonadaceae isolates. The numbers represent the mean inhibition zone diameters in mm for each antibiotic that was tested. The number of isolates for each cluster is divided between environmental (Env.) and animal (An.) isolates. The greyed-out fields represent those that are resistant according to the CLSI Susceptible Intermediate Resistant (SIR) status [ 24 ].
Abbreviations: GEN: gentamicin; TZP: piperacillin/tazobactam.
IPM: imipenem; MEM: meropenem; CIP: ciprofloxacin.

Dendrogram for Pseudomonadaceae that resulted from the cluster analysis. The y-axis (height) represents how close together observations were when they were merged into clusters. gower_distR refers to Gower distance which was used to calculate the distance matrix, and Ward's refers to the method used as the hierarchical clustering algorithm. The rectangular boxes represent each one of the four clusters (I, II, III, and IV).
4. Discussion
To fully understand and mitigate AMR, it is important to consider the role of the natural environment as part of the One Health approach that has been advocated towards this end. Wildlife species may be exposed to antibiotics and antimicrobial resistant organisms, and they may contribute to their dissemination. From a public health perspective, wildlife admitted to WRC have been mostly evaluated for their potential to carry and transmit zoonotic pathogens such as Salmonella spp. including raptors in Chile [ [31] , [32] , [33] , [34] ]; however, the role of WRC in the emergence and dissemination of AMR has been overlooked [ 35 ]. In this study, antibiotic resistant Gram-negative bacteria with reduced susceptibility to cefotaxime were characterized in both animal and environmental samples at a WRC in central Chile.
The results showed a high proportion of the cef-resistant bacterial subpopulation to also be resistant to three or more antibiotics (90% of animal isolates and 66.7% of environmental isolates). This finding is consistent with other studies that have also found remarkable percentages of resistant bacteria in wildlife undergoing rehabilitation. In one study, samples taken from injured wildlife admitted to a WRC in Spain revealed that 71% of all E. coli isolates recovered from animals were resistant to more than three individual antibiotics [ 36 ]. In a wildlife rescue center in Italy, resistance to 15/16 of antibiotics tested occurred among isolates from raptors and waterbirds, while there was resistance to 10/16 of antibiotics tested in isolates from passerine species [ 16 ]. Furthermore, another study found that 77.8% of northern elephant seals ( Mirounga angustirostris ) had antimicrobial resistant E. coli prior to release, compared to 38.4% of the seals at admission to a WRC [ 37 ]. These findings are compatible with others that have reported that wild animals either in captivity or closer to anthropogenic pressures tend to a higher prevalence of antibiotic resistant bacteria compared to those that are free-ranging or further from human influence [ [38] , [39] , [40] ].
Animals sampled in this study had been at the WRC an average of six months, and from those where retrieval information was known, they had been found near the city of Santiago, the capital of Chile, a large urban center. These two factors (time in captivity and proximity to human activities) could have a large influence on the antibiotic resistance outcome. In fact, there were two animals with a higher proportion of Enterobacteriaceae resistant isolates than others that had been at the WRC for a considerable time. One was a Patagonian land turtle ( C. chilensis ), admitted to the WRC after being confiscated from the illegal wildlife trade, that had spent over four months at the WRC and had received enrofloxacin treatment for 12 weeks. The prolonged antibiotic treatment and/or the prolonged captivity could have led to increased antibiotic resistance. The other case was an Andean fox cub ( L. culpaeus ) that had been at the WRC for almost 4 months and had not received antibiotic treatment. Age has been reported as a strong predictor of antibiotic resistance, with younger animals shedding a higher prevalence of resistant bacteria [ 41 , 42 ]. However, in a recent study evaluating antimicrobial resistance gene occurrence in Andean foxes, the authors did not find significant differences related to age [ 43 ].
There was no association between antibiotic treatment and frequency of resistant isolates. This could be explained by the small sample size ( n = 20), by a short time of exposure to the antibiotics, and by other factors that could not be accounted for in this study, such as location where the animal was originally found, as well as other components of the complexity of AMR epidemiology, and the wide presence of resistant bacteria in the built environment. Alternatively, the effect of the antibiotic therapy may have been short-lived, and the animals became repopulated with resident bacteria when the pressure of the antibiotics were off. This effect has been observed in other animal settings, with the duration of the effect being related to the fraction of the animal population that received antibiotic therapy [ 44 ].
Among the resistance patterns found in this study, it is important to highlight the identification of both Enterobacteriales and Pseudomonadaceae isolates resistant to carbapenems in the WRC environment and in the animals. This is very relevant from a public health perspective since these microorganisms were classified as critical priority by the World Health Organization (WHO) priority pathogens list for research and development of new antibiotics [ 45 ]. Another remarkable finding was the percentage (30.8%) of Enterobacteriales environmental isolates resistant to fosfomycin. This antibiotic with antibacterial activity against a wide range of gram-negative pathogens and some gram-positive pathogens, has been increasingly used worldwide in the last few years to treat uncomplicated urinary tract infections in humans when strains are resistant to other most commonly used drugs such as ciprofloxacin [ 46 , 47 ]. Antibiotic resistant bacteria in rehabilitated wildlife can be seeing from different perspectives. For instance, one aspect is the potential dispersal of antibiotic resistant bacteria from released wildlife to livestock and humans; another aspect is the environmental acquisition of antibiotic resistant bacteria by rescued wildlife, especially when this wildlife is found at or near urban areas or near livestock. However, a recent study conducted in the same geographical area as our study, found ESBL-producing E. coli in 24% of dogs, 3% of cows, but only in 0.5% of wildlife [ 48 ], values much lower than our results in the built environment. The different ways by which wildlife may play a role in the acquisition and in the dissemination of antibiotic resistant bacteria require further investigation.
A high percentage of Enterobacteriales (30.8% of the environmental isolates and 62.5% of the animal isolates) and Pseudomonadaceae (62.5% of environmental isolates and 33.3% of animal isolates) were resistant to ciprofloxacin, an antibiotic of the fluoroquinolone class. The wide use of enrofloxacin, another fluoroquinolone, at the WRC may have contributed to these results, as it has been noted in other studies [ 49 ]. In addition, commonly used disinfectants in hospital environments including this WRC such as quaternary ammoniums (QACs) could have contributed to an increase in fluoroquinolone resistant isolates. Even though there was no evidence to address this hypothesis at the genetic level in this study, there are documented interactions between the use of QACs and the emergence of fluoroquinolone resistance in bacteria [ 50 , 51 ].
In this study, the WRC built environment was an important reservoir of bacteria with reduced susceptibility to cephalosporins. While it was hypothesized that resistant bacteria from both types of samples would cluster together based on their antibiotic resistance patterns, the results did not support this hypothesis. Numerous antibiotic resistance profiles were observed in different bacterial species isolated here, indicating not only environmental contamination with a wide diversity of bacteria, but also a wide diversity of resistant bacteria in animals at the WRC. In our study, even though transfer of antibiotic resistant bacteria to WRC personnel was not investigated, we identified antibiotic resistant bacteria in human-touch surfaces, such as doorknobs, light switches, and areas within the WRC such as the reception and the kitchen. All these represent potential sites for dissemination of resistant bacteria to humans. Furthermore, the diversity of bacteria could be further analyzed using culture-independent methods, which would provide a broader perspective on the antibiotic resistance dynamics at the WRC and help overcome the inherent culture bias of culture-based methods [ 52 ].
The study design was cross-sectional, with samples only collected at one point in time. This means that results could have differed if samples had been collected at a different time. Furthermore, cross-sectional studies cannot provide an indication of the sequence of events, and thus it would not be possible to identify if the animals were admitted carrying resistant bacteria or instead they acquired the resistant bacteria during their stay at the WRC. Improved study designs consisting of longitudinal sampling of the animals from admission to their final outcome (release/euthanasia/transfer) would add valuable information about the potential emergence and/or acquisition of AMR at WRC.
In conclusion, an increased understanding on antibiotic use practices and AMR dynamics in wildlife rehabilitation is needed. It is critical to increase the knowledge about the influence of antibiotic and human exposure to wildlife populations, and when wild animals are placed in temporary captivity, to further understand the effects that hospitalization and reintroduction back into the natural environment can have on the potential emergence and spread of AMR, and thus on wildlife, human, and ecosystem health.
This work was supported by FONDECYT 1181167 and by the ANID Millennium Science Initiative/Millennium Initiative for Collaborative Research on Bacterial Resistance, MICROB-R, NCN17_81.
Credit author statement
Carla Baros Jorquera : Conceptualization, Methodology, Investigation, Writing; Andrea I. Moreno-Switt : Conceptualization, Methodology, Investigation, Writing, Supervision, Funding acquisition; Nicole Sallaberry-Pincheira : Resources, Conceptualization, Writing-Review & Editing; Jose M. Munita : Writing-Review & Editing, Funding acquisition; Camila Flores Navarro : Writing-Review & Editing; Rodolfo Tardone : Methodology, Writing-Review & Editing; Gerardo González-Rocha : Methodology, Writing-Review & Editing; Randall S. Singer : Methodology, Writing- Review & Editing; Irene Bueno: Methodology, Formal analysis, Writing, Supervision.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
We thank all the staff and volunteers at the UFAS UNAB/Buin Zoo wildlife rehabilitation center at the Universidad Andrés Bello (Santiago, Chile) that helped to make this study possible.
Appendix A Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2021.100298 .
Appendix A. Supplementary data
Clinical and Laboratory Standards Institute (CLSI) breakpoints for the isolates included in the study.

IMAGES
COMMENTS
A thesis submitted to Johns Hopkins University in conformity with the requirements for the degree of Master of Arts Baltimore, Maryland August 2020 . ii Abstract ... Antimicrobial resistance is a ticking time bomb not only for the UK but also for the rest of the world. We need to work with everyone to ensure the apocalyptic scenario of widespread
Shlaes, et al. 1997), particularly in view of increasing drug resistance and diminishing antibiotic pipelines (Spellberg, et al. 2008). In addition to the emergence of resistance, antibiotic use was estimated to be responsible for about 142,505 visits to emergency rooms in the United States for drug-related adverse events attributable to systemic
2. Antimicrobial Resistance: A Worldwide Public Health Emergency. In a recent estimate, about 3.57 million of 4.95 million deaths worldwide were associated with antimicrobial resistance in 2019 [], higher than many other well-known causes of mortality, including malaria and HIV/AIDS.According to the study, the worldwide burden of AMR is significantly higher than the 700,000 deaths per year ...
In Asia, the Japanese Veterinary Antimicrobial Resistance Monitoring system was initiated in 1999, and the Korean Nationwide Surveillance of Antimicrobial Resistance was established in 1997 in South Korea . In Indian context, there are no regulations for the use of antibiotics in food animals. The Global Antibiotic Resistance Partnership (GARP ...
Innately, soil bacteria produce antimicrobial substances to. inhibit their competitors' growth, and pristine locations that are isolated from human. contact display very small levels of antibiotic resistance (9, 10). Furthermore, the. production of some of these substances are only expressed when the bacteria receive.
1. Introduction. Antimicrobial resistance (AMR) has been recognised as one of the greatest threats to human health over the last decade [1,2,3,4,5].The World Health Organization (WHO) have estimated that antimicrobials add approximately 20 years to life expectancy globally, but this advantage is diminishing due to AMR [].In 2019 there were an estimated 4.95 million deaths globally due to AMR [].
Antibiotic resistance is a serious public health problem. Assessment of knowledge, attitude and practices of antibiotic use in university students can greatly impact how best to tackle the growing threat of the antibiotic resistance and its related issues [18, 20]. In the present study, students in health related majors had better knowledge ...
Bellarmine University Honors Thesis 29 April 2020 Advisor: Dr. Amanda Krzysiak Readers: Dr. Steven Wilt and Dr. Savita Chaurasia ... Antibiotic resistance infections can be acquired through two methods: contraction of antibiotic resistant bacteria from an outside source or antibiotic resistance occurring due to the
UNEXPECTED ANTIBIOTIC RESISTANCE IN GRAM-POSITIVE BACTERIA RECOVERED FROM THE CHESAPEAKE BAY AND ASSOCIATED RIVERS AND INVESTIGATING THE ANTIMICROBIAL ACTIVITY OF A WIDE VARIETY OF ESSENTIAL OILS AS A MEANS TO IDENTIFY NOVEL DRUG TARGETS by Jennifer Leigh Rivers A thesis submitted to the Bloomberg School of Public Health at the Johns Hopkins
The emergence of antimicrobial resistance (AMR) results in drug inefficiency and persistent infections, with a subsequent increase in the risk of severe disease and transmission. Antibiotic ...
Introduction. Antimicrobial resistance (AMR) is a major global health challenge, causing substantial morbidity and death globally. Understanding the molecular mechanisms that underlie resistance ...
Antimicrobial resistance (AMR) is among the top ten global public health threats, demanding immediate attention and action [ 1 ]. To effectively tackle AMR, it is crucial to have a comprehensive understanding of the extent of the problem. Specifically, reliable, accurate, and representative information on the incidence of resistant infections ...
Hayat K, Jamshed S, Rosenthal M, et al. Understanding of pharmacy students towards antibiotic use, antibiotic resistance and antibiotic stewardship programs: a cross-sectional study from Punjab, Pakistan. Antibiotics. 2021;10(1):66.
Despite the recent passage of an antimicrobial stewardship programs (ASP) mandate in California and the Centers for Disease Control and Prevention (CDC) guidelines for a minimum standard ASP in hospitals, literature on the impact of ASPs on antimicrobial resistance (AMR) rates in hospitals is sparse.
Antimicrobial resistance phenotypes of E. coli isolates. ... The highest content of tet genes in poultry litter isolates confirms the thesis put forward by Furtula et al. 44, ...
Bloodstream infections (BSI) pose a significant health challenge, particularly among children. These infections in the pediatric population are linked to undesirable levels of morbidity and mortality, highlighting the urgent necessity for robust diagnostic and therapeutic approaches [].Adding complexity to this challenge is the concerning trend of increasing antimicrobial resistance among the ...
Antimicrobial resistance (AMR) is widely acknowledged as a global problem, yet in many parts of the world its magnitude is still not well understood. This review, using a public health focused approach, aimed to understand and describe the current status of AMR in Africa in relation to common causes of infections and drugs recommended in WHO treatment guidelines.
Nearly 150 million cases of urinary tract infections (UTIs) are reported each year, of which uncomplicated cystitis triggers > 25% of outpatient prescriptions of oral antimicrobial treatment (OAT). OAT aids immune cells infiltrating the urothelium in eliminating uropathogens capable of invading the urothelium and surviving hyperosmotic urine. This self-evident adaptability of uropathogens and ...
eRepository @ Seton Hall
Antimicrobial resistance (AMR) is increasingly being recognized as a serious global health threat, as demonstrated by high-level policy initiatives, such as the Transatlantic Taskforce on Antimicrobial Resistance (TATFAR), the Global Antibiotic Resistance Partnership (GARP), the Global Health Security Agenda (GHSA), the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR), and the ...
China, the leading global producer and consumer of antibiotics, faces growing antimicrobial resistance as a result of inappropriate use of antibiotics in clinical, agricultural, livestock, and aquacultural settings (1, 2). China's first national action plan to contain antimicrobial resistance yielded positive outcomes, but the prevalence of clinical multidrug-resistant bacteria in China ...
Background Antimicrobial resistance is an important global health challenge. The current study aimed to assess the level of awareness and knowledge of antimicrobial resistance and factors associated with knowledge among adults in Dessie City, Ethiopia. Methods A community-based cross-sectional study was conducted among 407 adults in Dessie City from June to July 2021. A systematic random ...
A report on antimicrobial resistance surveillance in Europe showed that the resistance rate of K. pneumoniae against carbapenems was 6.1% in 2016, without a significant change from 2013 to 2016. The prevalence of CRKP in Greece was the highest, up to 61.9% . In summary, just like the global trend, Gram-negative bacterial resistance in China is ...
While decay rates of total fecal bacterial indicators in soil have been frequently reported, very few studies have been completed comparing persistence of the antibiotic-resistant counterparts. Additionally, little is known about how the multi-drug resistance of ARB changes over time after biosolids amendment or TWE irrigation.
WHO has published its first global research agenda for the world's scientists to address the most urgent human health priorities to combat antimicrobial resistance (AMR). It outlines 40 research topics on drug-resistant bacteria, fungi and Mycobacterium tuberculosis that must be answered by 2030, in line with the Sustainable Development Goals.. The WHO Global Research Agenda for AMR in human ...
In Turkey, the rate of this resistance in separate studies had increased from 32.7% to 50% in 2009 and 2013.[24,48] In a Spanish study, E. coli antibiotic resistance at 2005, 2009, and 2011 was examined; the results showed that resistance to amoxicillin had increased from 4.5% in 2005 to 55.6% in 2011.
Antibiotic resistance is a global public health problem that is responsible for increased patient morbidity and mortality and financial burden. Dental antibiotic prescribing contributes to approximately 10% of all antibiotic prescriptions, and an estimated 80% of that prescribing is deemed inappropriate. Dental antimicrobial stewardship (AMS) has an important role to play in international ...
Synanthropic filth flies are common where sanitation is poor and fecal wastes are accessible to them. These flies have been proposed as mechanical vectors for the localized transport of fecal microbes including antimicrobial resistant (AMR) organisms and associated antimicrobial resistance genes (ARGs), increasing exposure risks.
1. Introduction. The interconnectedness between humans, animals, and the natural environment (otherwise known as One Health) is key in understanding and mitigating antimicrobial resistance (AMR) given that resistant bacteria and resistance genes have the ability to move between these three compartments [, , ].Of these three compartments, the role of the natural environment (e.g., soil, water ...